Simulation-based calibration of Bayesian NBBP model
sbc.RmdTheory
One sanity check of a Bayesian model implementation is simulation-based calibration. We simulate from a model by drawing a dataset from the prior predictive distribution. Then we fit the model and check whether the true value was covered at many quantiles. If we repeat this process enough times, the percentage of true values covered at the th percentile should be . That is, when we draw from the prior predictive, the posterior should obey frequentist coverage properties.
We can also examine how well point estimates (say, the posterior mean) recapitulate the truth along the way.
This is a best-case sort of test, the results of which come with the package as data.
Main results
There are two sets of coverage results includes as package data in
sbc_quants. One is for completely-observed chains, and
consists of analyses of 1000 datasets of 25 chains each. The other is
for the same datasets, but with only 50% of the cases observed. (The
true per-case detection probability was supplied to the model.) In both
cases, the nominal and observed 1st through 99th percentiles of coverage
of
and
are reported.
Here are the results for :
library(nbbp)
library(ggplot2)
data(sbc_quants)
sbc_quants |>
ggplot(aes(x = quantile, y = r_covered)) +
facet_wrap(~observations) +
theme_minimal() +
geom_point() +
geom_abline(intercept = 0, slope = 1, col = "red") +
xlab("Quantile") +
ylab("Observed coverage of R at quantile")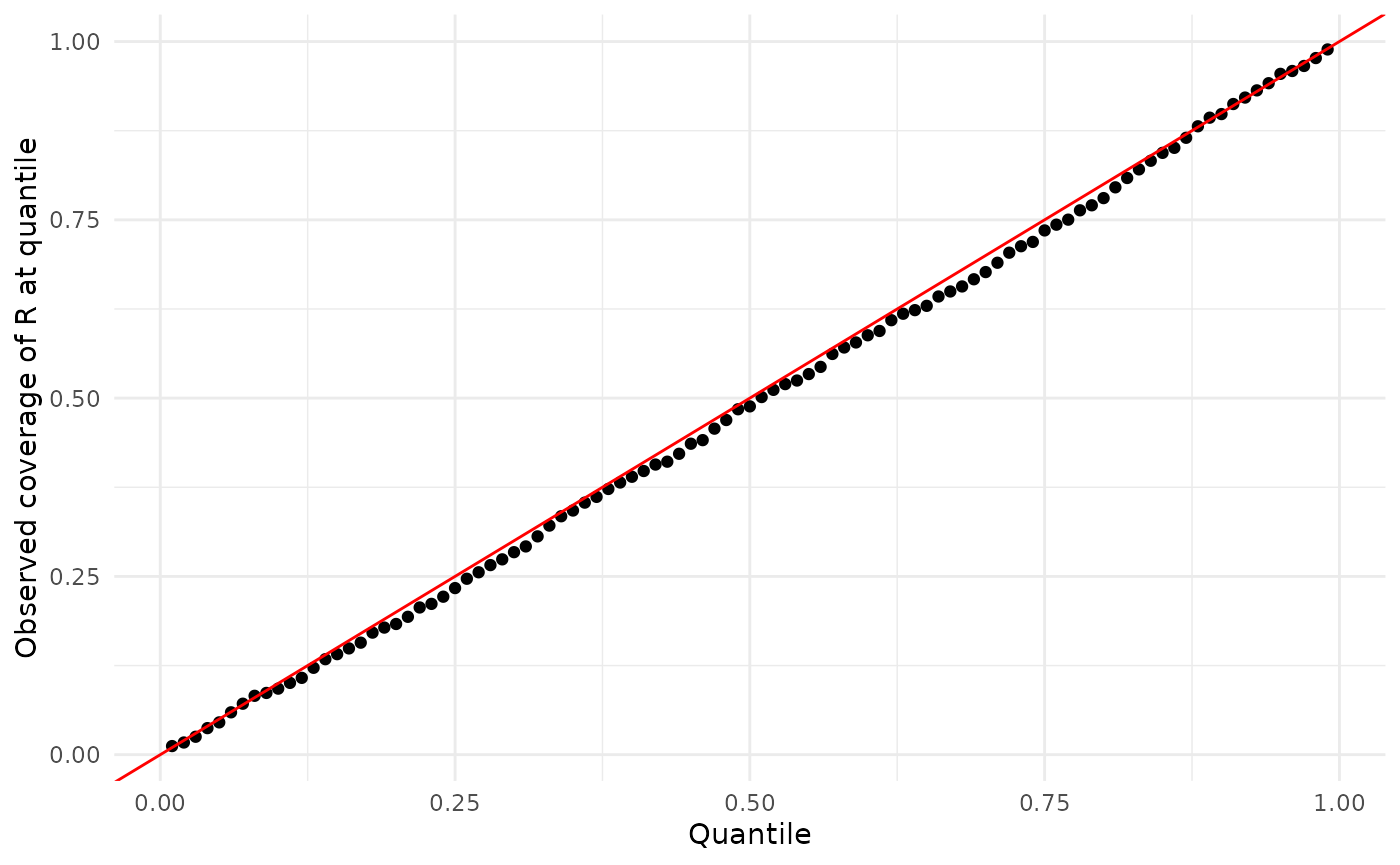
Here are the results for :
library(ggplot2)
data(sbc_quants)
sbc_quants |>
ggplot(aes(x = quantile, y = k_covered)) +
facet_wrap(~observations) +
theme_minimal() +
geom_point() +
geom_abline(intercept = 0, slope = 1, col = "red") +
xlab("Quantile") +
ylab("Observed coverage of k at quantile")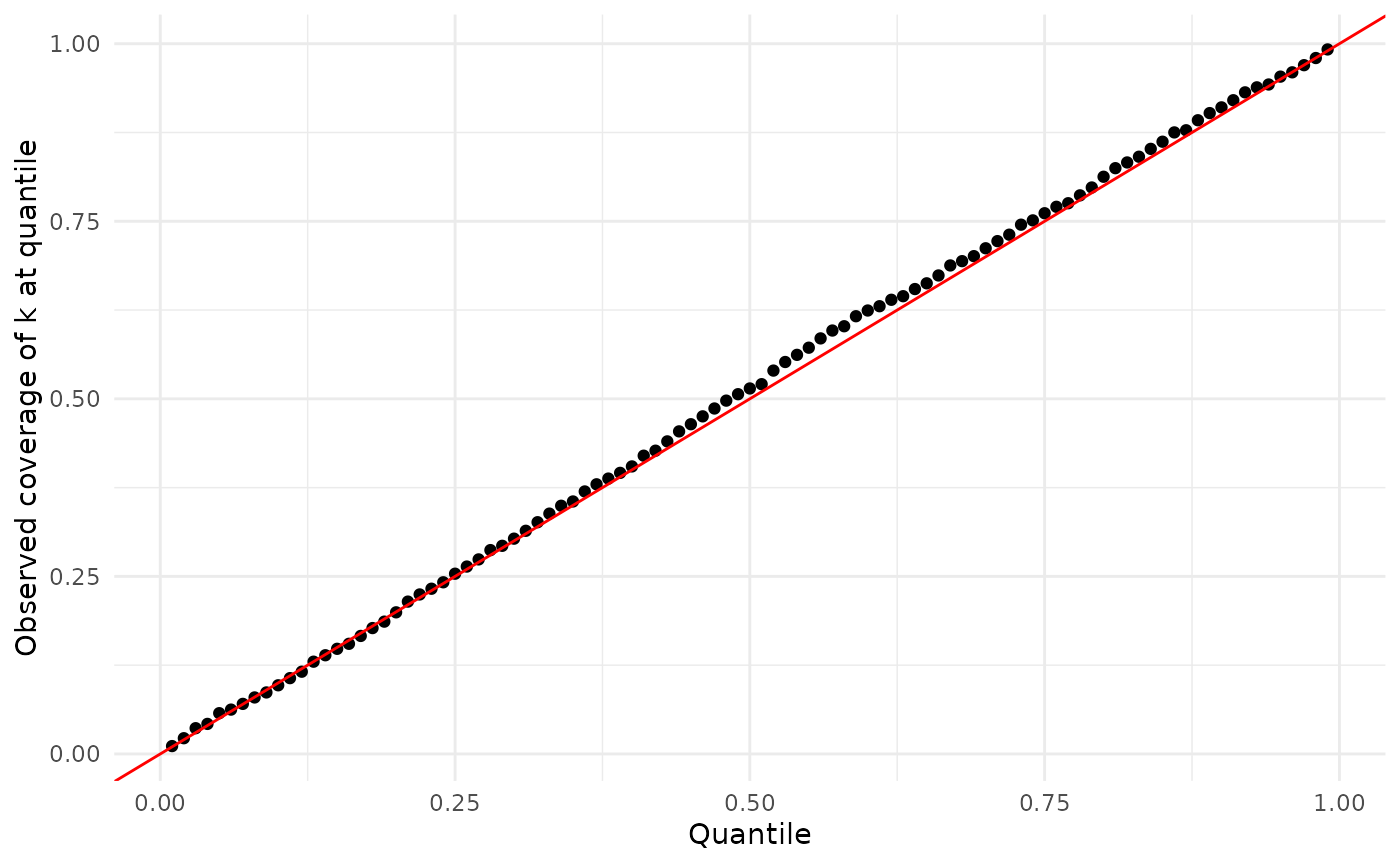
Ancillary results
Also available (as data(sbc_ests)) is information about
posterior and MCMC quality, for both the completely- and
partially-observed simulations.
Estimating
Here we plot the posterior median against the true simulating , with the 95% CI as a bar. Points are red (with non-transparent bars) when the true value is not in the 95% CI, and blue (with transparent bars) otherwise.
data(sbc_ests)
sbc_ests |>
dplyr::mutate(covered = dplyr::case_when(
r_true >= r_low & r_true <= r_high ~ TRUE,
.default = FALSE
)) |>
ggplot(aes(
x = r_true, y = r_point,
ymin = r_low, ymax = r_high,
col = covered,
)) +
facet_wrap(~observations) +
theme_minimal() +
geom_abline(intercept = 0, slope = 1, col = "grey", lty = 2, lwd = 2) +
geom_errorbar(aes(alpha = 1 - covered), show.legend = FALSE) +
geom_point() +
xlab("true R") +
ylab("estimated R") +
scale_x_continuous(trans = "log") +
scale_y_continuous(trans = "log")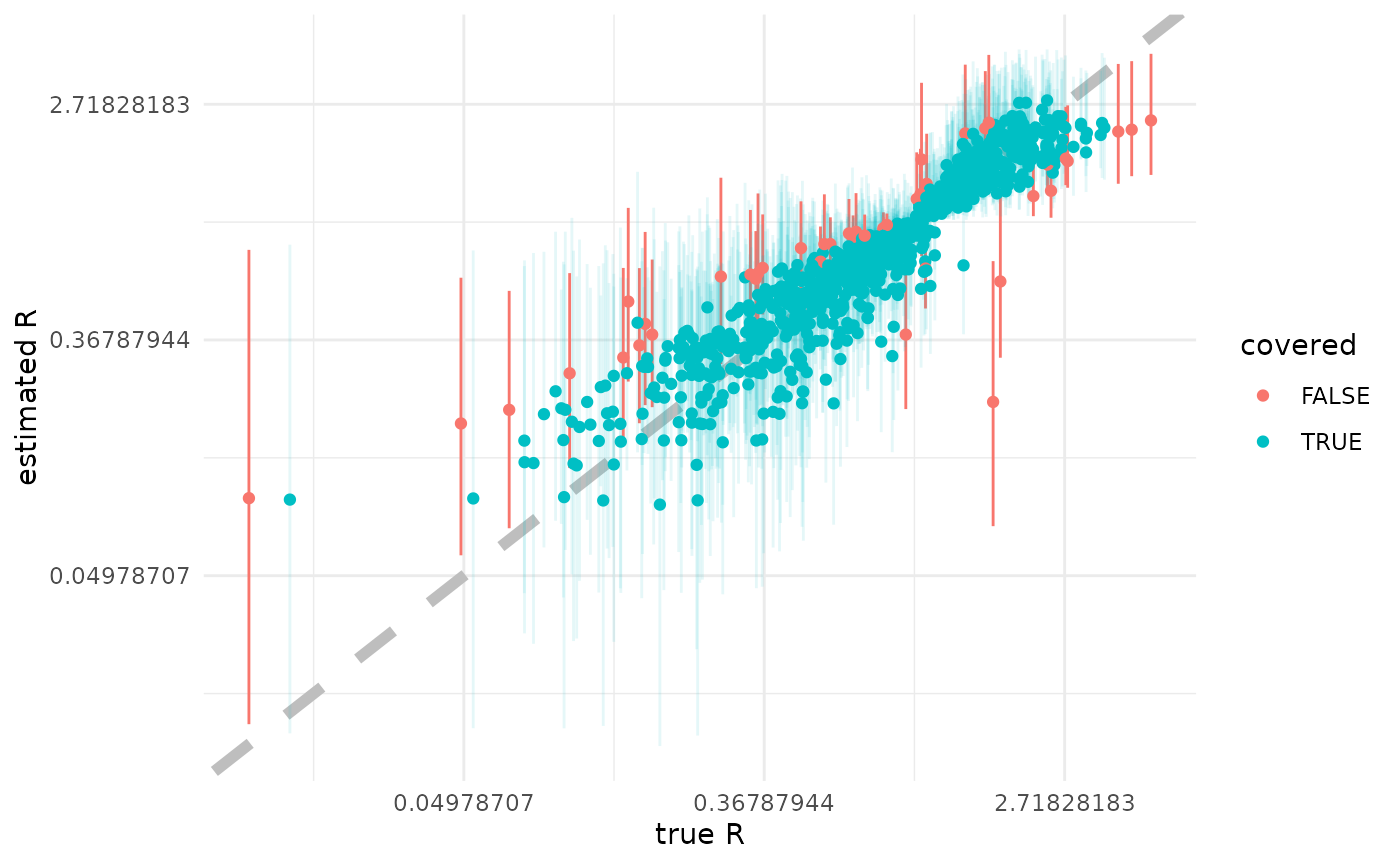
Estimating
Here we plot the posterior median against the true simulating , with the 95% CI as a bar. Points are red (with non-transparent bars) when the true value is not in the 95% CI, and blue (with transparent bars) otherwise.
data(sbc_ests)
sbc_ests |>
dplyr::mutate(covered = dplyr::case_when(
k_true >= k_low & k_true <= k_high ~ TRUE,
.default = FALSE
)) |>
ggplot(aes(
x = k_true, y = k_point,
ymin = k_low, ymax = k_high,
col = covered,
)) +
facet_wrap(~observations) +
theme_minimal() +
geom_abline(intercept = 0, slope = 1, col = "grey", lty = 2, lwd = 2) +
geom_errorbar(aes(alpha = 1 - covered), show.legend = FALSE) +
geom_point() +
xlab("true k") +
ylab("estimated k") +
scale_x_continuous(trans = "log") +
scale_y_continuous(trans = "log")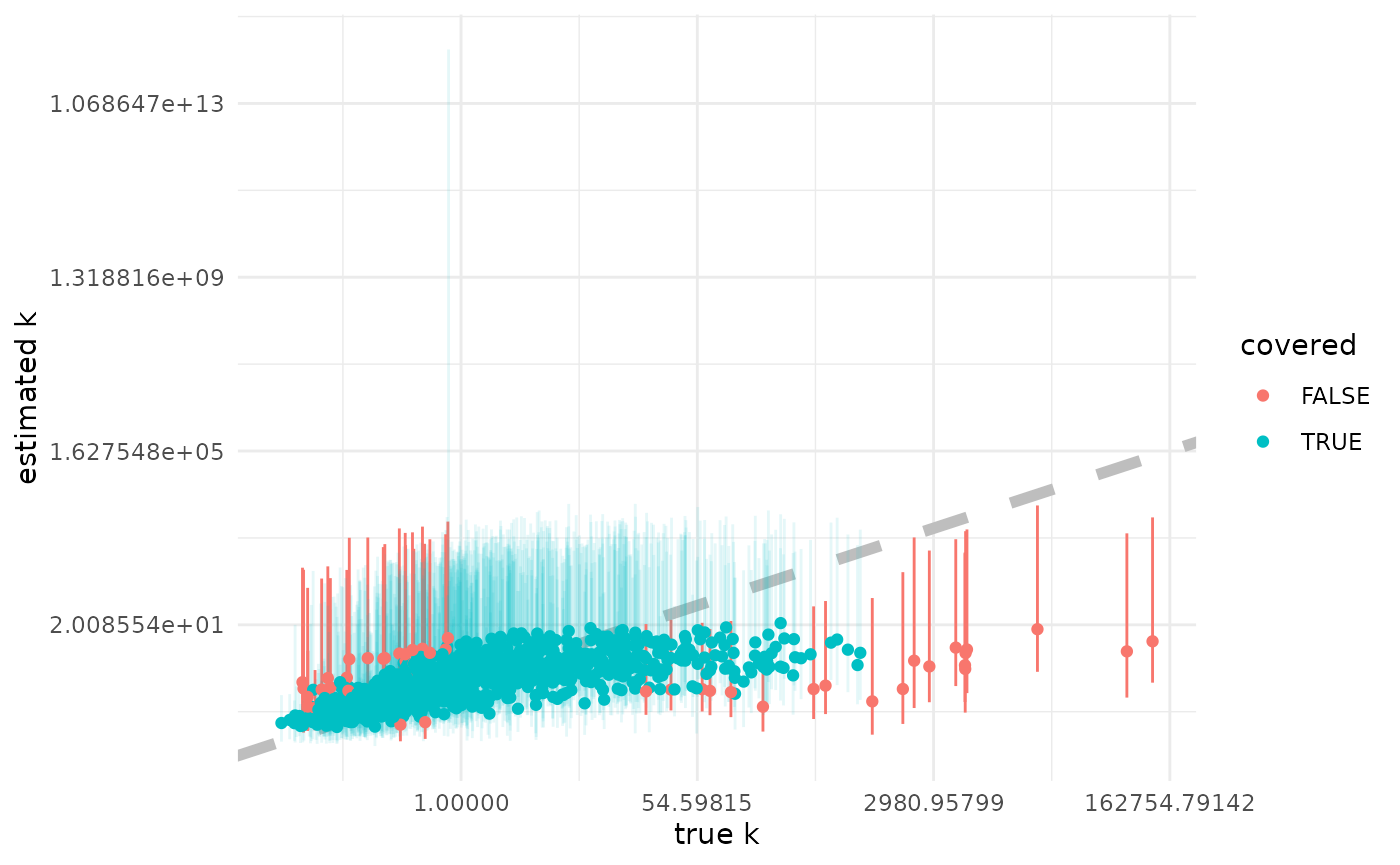
MCMC behavior
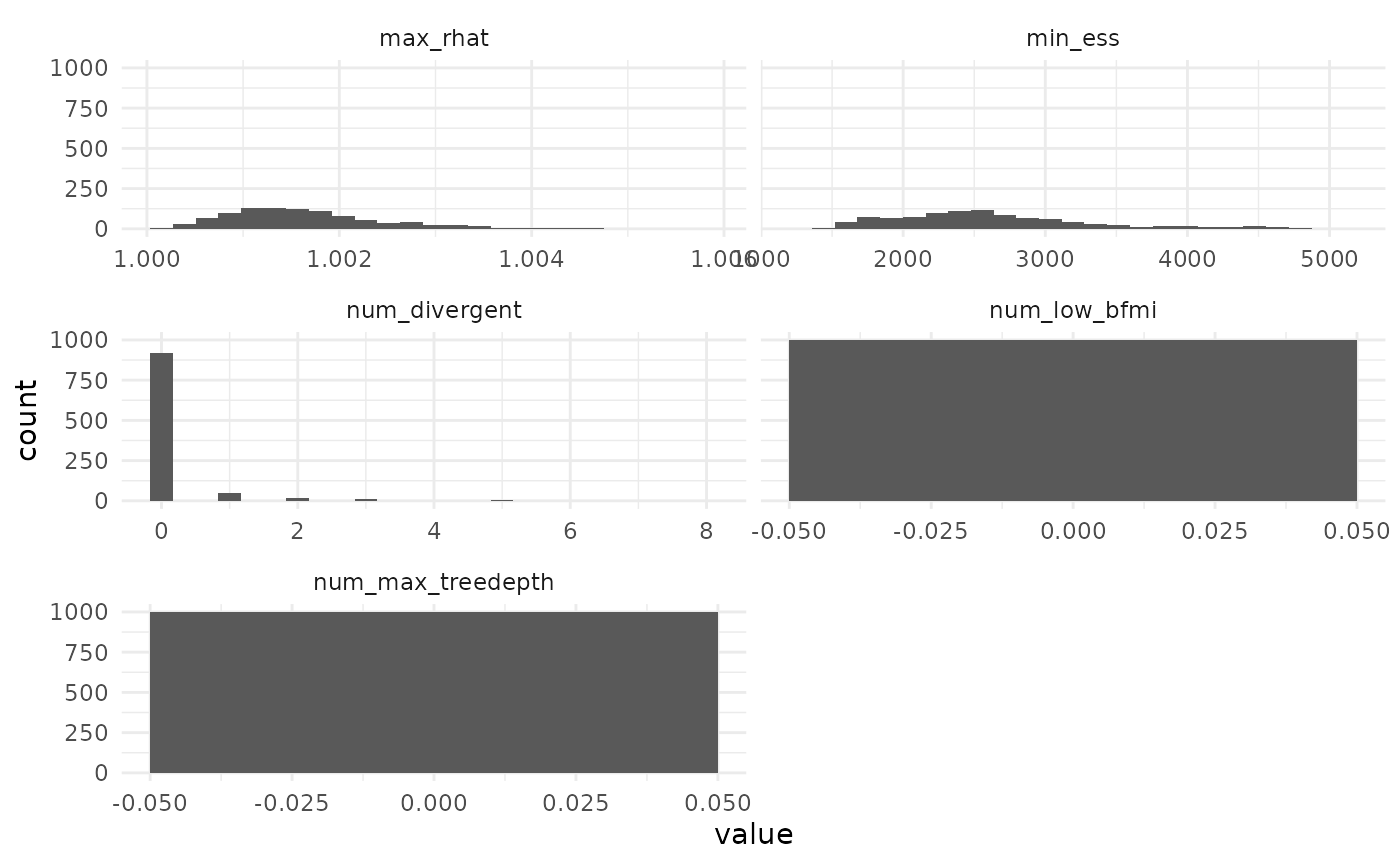
We can also see that most MCMC performed well, with a few exceptions that are visible mostly through the limits chosen for the axes when we plot all the diagnostics.
sbc_ests |>
dplyr::mutate(index = dplyr::row_number()) |>
tidyr::pivot_longer(
cols = c("min_ess", "max_rhat", "num_low_bfmi", "num_divergent", "num_max_treedepth"),
names_to = "name",
values_to = "value"
) |>
ggplot(aes(x = value)) +
theme_minimal() +
geom_histogram(bins = 25) +
facet_grid(observations ~ name, scales = "free_x")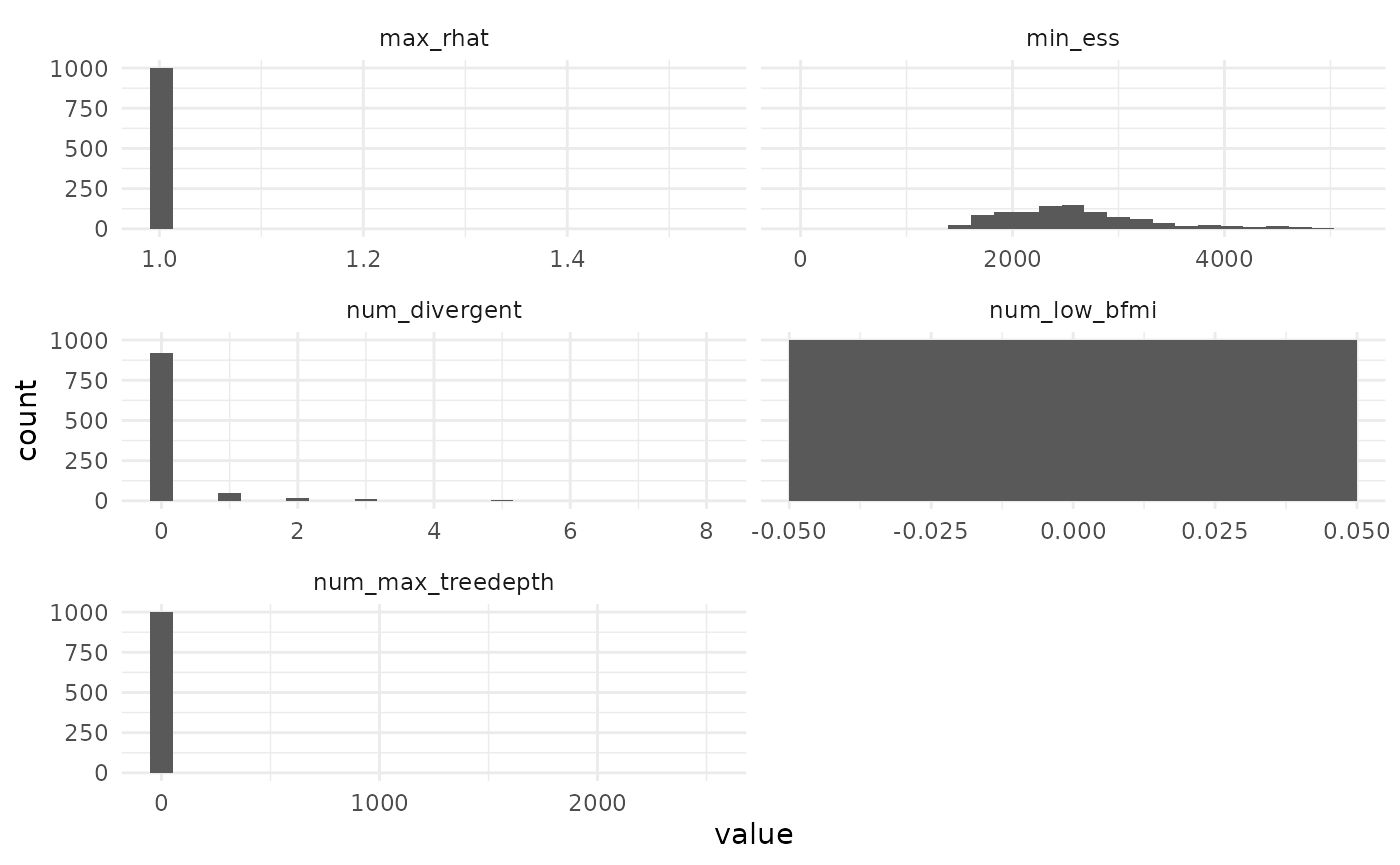
Filtering for large n_eff and small Rhat,
as was done to obtain the quantiles in sbc_quants removes
the runs with pathologically bad num_max_treedepth as
well.
sbc_ests |>
dplyr::filter(min_ess > 1000 | max_rhat < 1.005) |>
dplyr::mutate(index = dplyr::row_number()) |>
tidyr::pivot_longer(
cols = c("min_ess", "max_rhat", "num_low_bfmi", "num_divergent", "num_max_treedepth"),
names_to = "name",
values_to = "value"
) |>
ggplot(aes(x = value)) +
theme_minimal() +
geom_histogram(bins = 25) +
facet_grid(observations ~ name, scales = "free_x")