Default priors
default_priors.RmdA Gamma prior is placed on
,
and a HalfNormal on
.
The user may specify hyperparameters of the Gamma distribution (its
shape and rate) and the HalfNormal (its scale parameter). Samples from
the default prior are available in the package data object
prior_predictive (created by this
script), which we will use to visualize the priors.
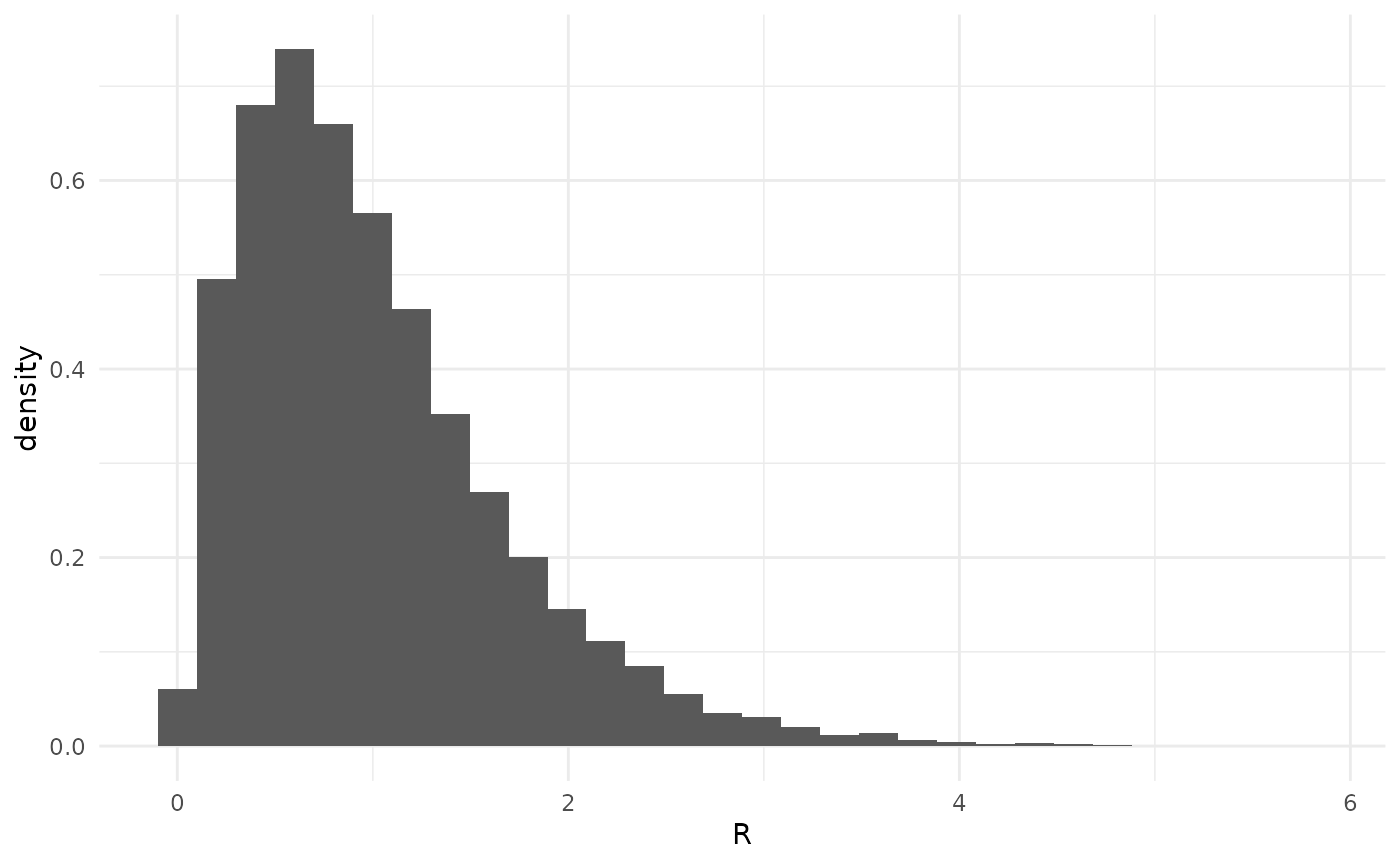
Default prior on
The parameter is the average number of infections caused by one infected individual (that is, it is the reproduction number).
The default prior on is Gamma(shape = 2.183089, rate = 2.183089). This has a mean of 1 and stipulates that is most likely between 1/2 and 2 (with 66.67% of prior mass in this region). The choice of mean 1 serves to provide some prior regularization (pull) towards 1. In regimes where is small, this should make estimation somewhat conservative, erring on the side of larger . The tradeoff is that in regimes where is large, we can under-estimate . (For simulation-based testing of such performance, see the vignettes “Simulation-based testing” and “Simulation-based calibration.”)
Some might wonder why the prior on is not more informative, while others might wonder why it is not less informative. Broadly, the prior is intended to be slightly informative while simultaneously allowing the data to express themselves in regions of which may be encountered during expected usage of this package. Inference of from final size data under a negative binomial branching process model is strongly associated with the “stuttering chains” regime of non-sustained transmission. This is typically a subcritical () regime, though with sufficient overdispersion (see below for the effect of ) is is possible in the supercritical regime.
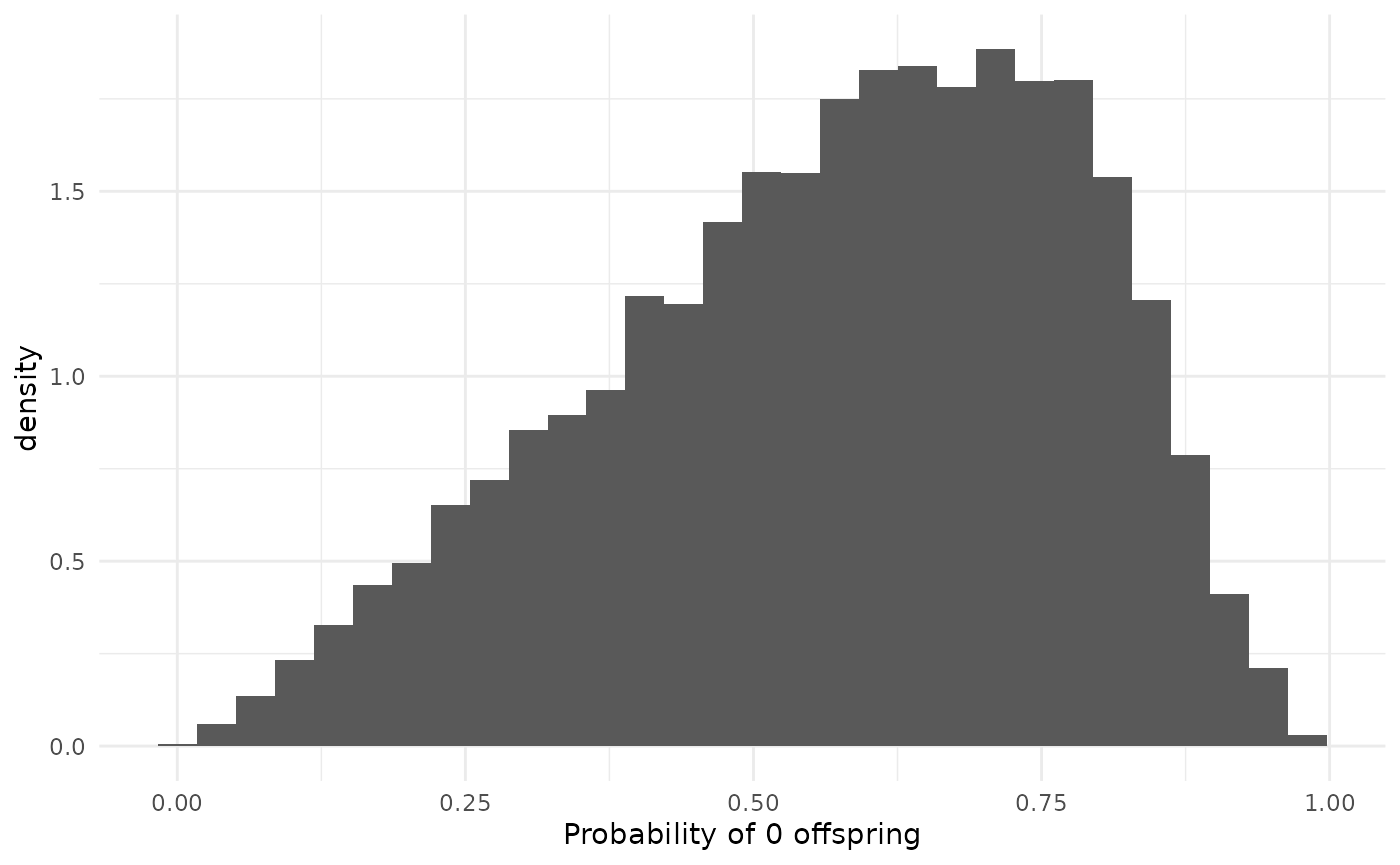
Default prior on
Note that the prior on is implicit (though we can still discuss it); the explicit prior is placed on . The choice of a HalfNormal on follows best-practices for a “weakly” informative prior on the NegativeBinomial over-dispersion parameter.
The parameter controls the dispersion of the offspring distribution, as well as the final size distribution, and plays a strong role in the probability of extinction of a chain. When is large, there is less dispersion relative to the mean, and in the limit of , the NegativeBinomial approaches the Poisson. At , the NegativeBinomial coincides with the Geometric distribution. When is small, the variance of the offspring distribution gets larger, increasing the variance of the chain size distribution. This increase in variance also increases the probability that an individual has no offspring, increasing the extinction probability. In the limit as , all chains die out after the index case and there is no information about whatsoever.
Since both and affect the extinction probability, our inference of one affects our inference of the other. Did the observed chains go extinct because most infections cause, on average, few new infections, or because many infections cause none at all, while others cause many? The prior pulls towards larger , which, handwavily, means the model should prefer to answer this question in terms of the mean.
The default prior has a median of 1, placing 50% of the prior mass in both the strongly-overdispersed regime and the less-overdispersed regime.
The (implicit) default prior on
has heavy tails, reflecting the prior’s pull towards the Poisson regime
of
.
Truncating to
for visualization purposes, the (conditional implicit) prior is
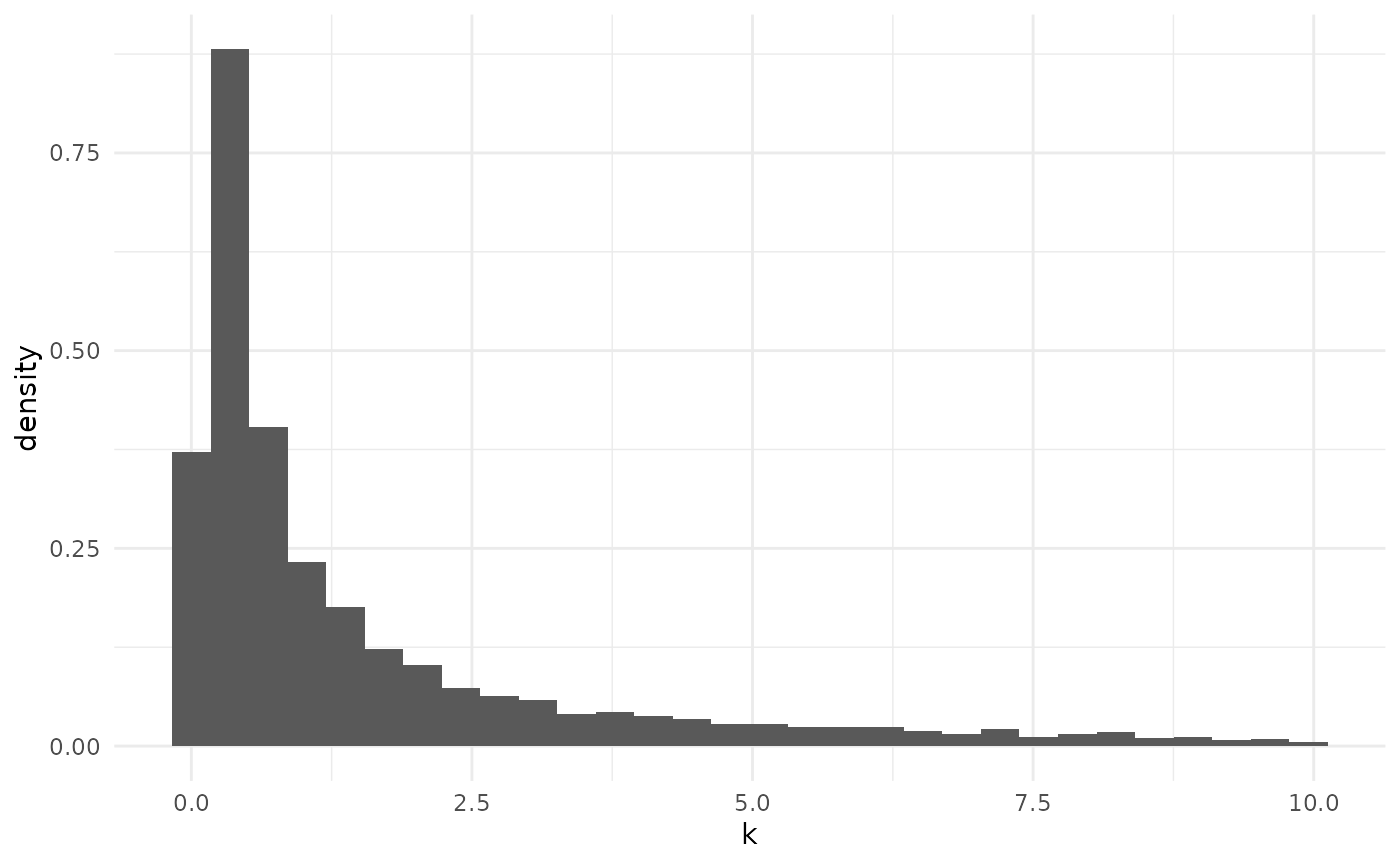
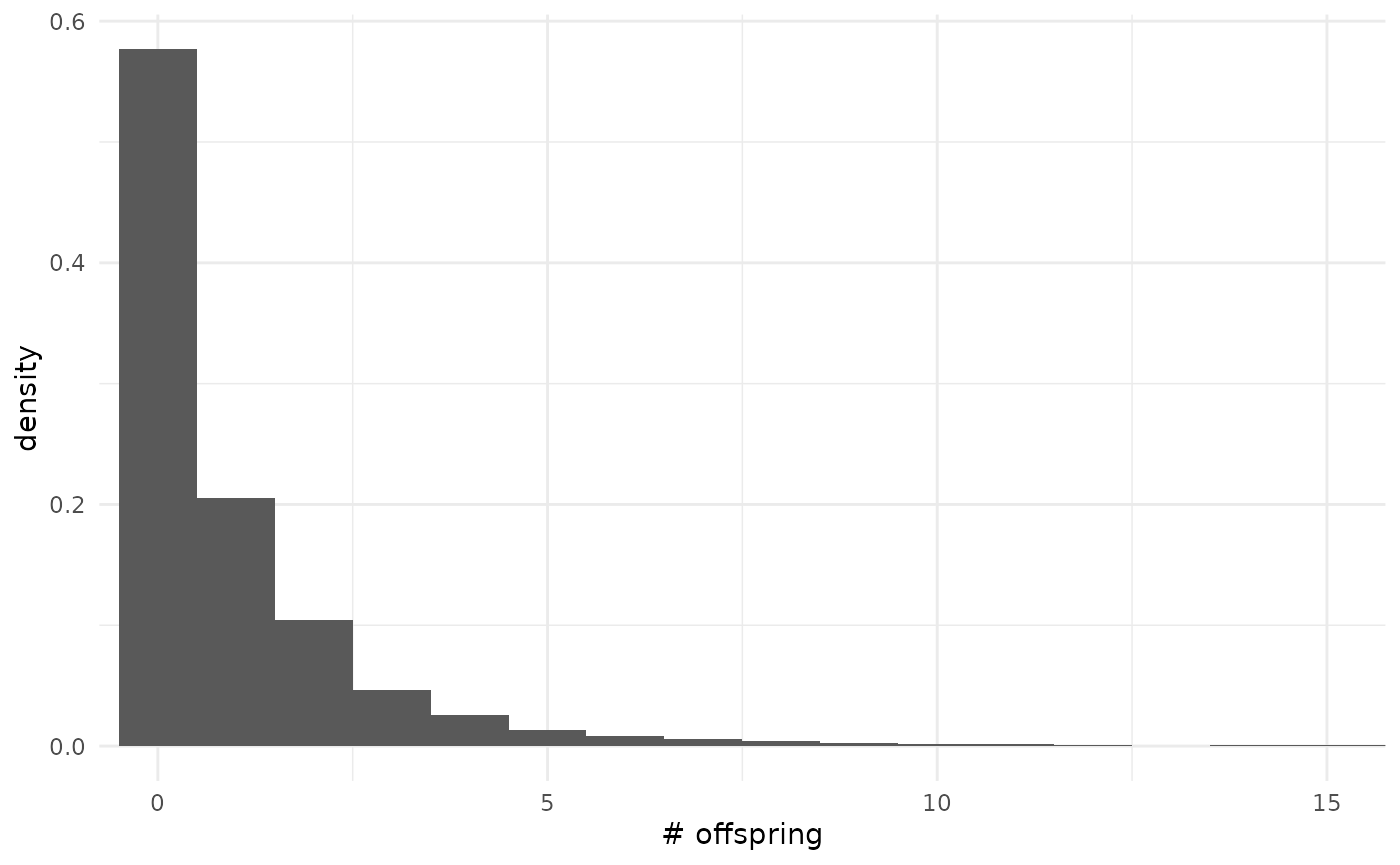
Default prior predictive offspring distribution
Conditioned on
and
,
the offspring distribution is negative binomial. By marginalizing this
conditional distribution across the priors on
and
,
we obtain the prior predictive offspring distribution.
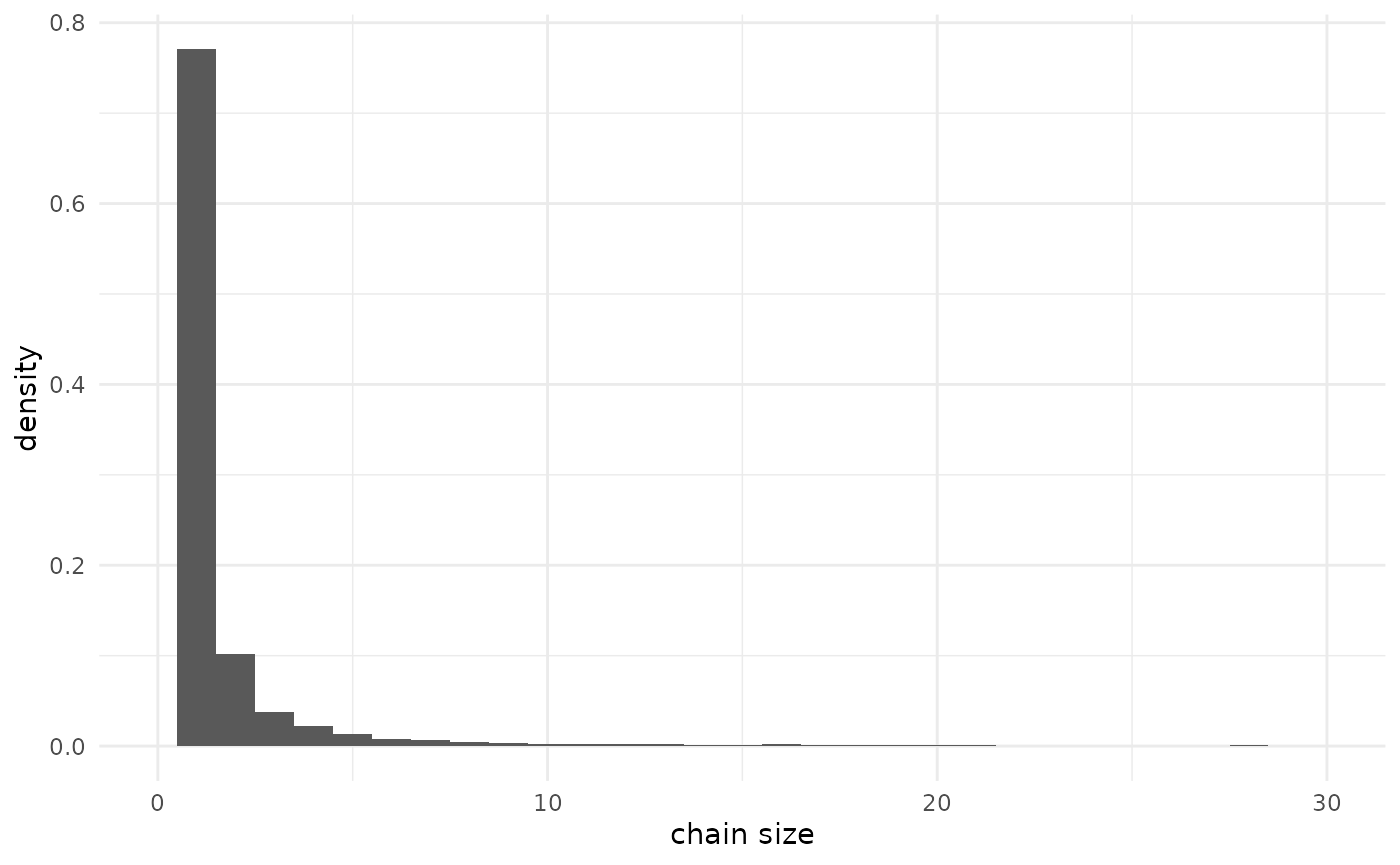
Default prior predictive chain size distribution
The prior predictive distribution on chain sizes has two components.
There is a 14% chance of a non-extinct (infinite) chain (see the
vignette “Advanced data” for more on this). Conditional on extinction
(finite size), there are long tails, with a 14% chance of a chain size
above 100. Focusing in on chains of 100 or fewer for visualization, the
(marginal with respect to
and
,
conditional with respect to the chain size) prior distribution is: