Implementation details
details.RmdConditioning and multimodality of the likelihood surface
As can be seen in the vignette “nbbp,” the likelihood surface sometimes has a mode at 0. It can also be multimodal when conditioning is applied.
Conditioning on observation sizes
In particular, multimodality can be induced when conditioning on observing chains of at least some size. An example of this can be seen using the pneumonic plague data considered in “Advanced data” where the likelihood must be conditioned on observing at least two cases to match the data-collection process.
library(nbbp)
library(ggplot2)
data(pneumonic_plague)
pneumonic_plague[4] <- Inf
lnl_surface <- compute_likelihood_surface(
pneumonic_plague,
condition_geq = rep(2, 19),
r_grid = seq(1.0, 1.35, length.out = 100),
k_grid = exp(seq(log(0.01), log(9999), length.out = 100))
)
lnl_surface |>
dplyr::mutate(
log_dens = ifelse(log_dens >= max(log_dens) - 1, log_dens, NA)
) |>
ggplot(ggplot2::aes(x = r, y = k, fill = log_dens)) +
theme_minimal() +
geom_tile() +
scale_y_continuous(transform = "log") +
scale_fill_viridis_c(option = "magma")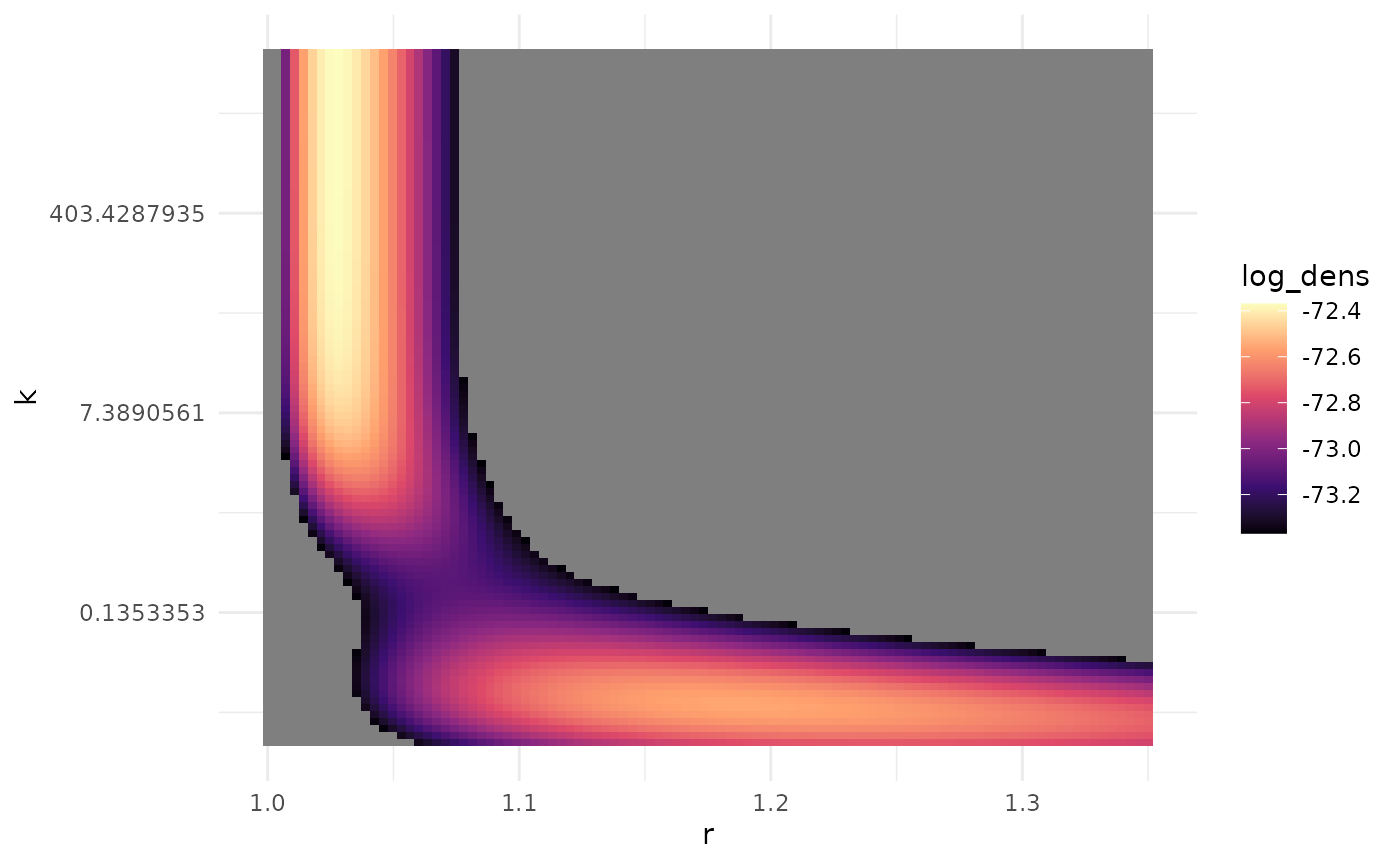
Inferring an unconstrained
Some applications of the negative binomial branching process model
have conditioned the likelihood on extinction to study the
case. In this way, it can be guaranteed that all chain sizes are finite,
even when infinite chains are possible. However, Waxman and Nouvellet
(2019) show that when conditioning there are two indistinguishable
modes, one with
and one with
.
For this reason, we treat non-extinct chains separately in both
stan code for inference and R code for
simulation. In inference, a non-finite chain provides guarantees that
,
this is the necessary condition for a negative binomial branching
process to never go extinct. Therefore, the presence or absence of such
chains is itself informative (see the vignette “Advanced data” for an
application). In simulation, this means that when
,
some samples may be Inf, indicating a non-extinct
chain.
Mathematically, we partition the model outcomes into finite and infinite chains. That is, a chain either goes extinct (has finite size) or doesn’t, and this happens according to the extinction probability . Formally, the possible chain sizes are . We can write the likelihood for one chain size as where the chain size which can take finite values as well as the value “”, is the indicator function, and () is the event that the chain goes (does not go) extinct. For conditionally finite chains, we can write Using this expression reduces the likelihood of chain size to
Numerical approximations
Probability of extinction
Except for very specific values of
,
the extinction probability is not available in closed form. In
stan, we use Newton’s method to obtain a numerical solution
when it is needed.
CDF
While the PMF for the chain-size distribution is available in closed form, there is no closed form solution for the CDF.
Compromises for efficiency
For efficiency, when drawing random chain sizes the package uses
base::sample() on a pre-computed, truncated, set of values.
This enables rapidly sampling many realizations for a pair of
values. Truncation is hard-coded at 1,000,000 which in practice appears
to cover at least 99.999% of the probability mass. The most mass is
unaccounted for when
and
.
Censoring
When brute-force computing the CDF (by summing the PMF), sometimes it
evaluates to numbers slightly greater than 1. In practice, this appears
to happen exclusively for
and the maximum seems to be less than
.
This is noticeable when censoring observations, where for chain size
we need
.
When the CDF exceeds one, this is negative and MCMC cannot appropriately
explore the
region. To avoid these issues, the stan model code checks
that the CDF does not exceed 1 over the range it needs to be evaluated.
If it ever exceeds 1, we rescale so that the maximum CDF is 1.0 - machine_precision().
Incompletely observed chains
Incompletely-observed chains require either inferring the unobserved true size via data augmentation or marginalizing the likelihood over the true size. We adopt the latter approach, which is implemented for a binomial sampling model of observed chain sizes as discussed in the “Advanced data” vignette. We must therefore approximate the infinite sum over the unobserved true size given the observed size and a per-case observation probability :
In practice, we must evalutate the finite sum up to some , leaving an error,
Under the hood, nbbp uses a dynamic approximation to
choose
such that the absolue error is no larger than a pre-specified
.
As we show below, by choosing
in terms of the quantile of a negative binomial distribution, we can
achieve this bound.
The bound can most readily be understood in two parts. First, and introducing a later-convenient factor of ,
Rewriting the RHS,
This can be recognized as the upper tail probability of a negative
binomial distribution, in R’s parameterization, with prob
parameter
and size parameter
,
evaluated at
.
Thus, and using the more common lower tail, we can write
Ergo, if we choose to set
to the
quantile of said negative binomial, we will keep the error no larger
than
.
That is,
In practice, this bound appears quite lenient. Numerical experiments suggest that, at default package settings for selecting , the error is perhaps roughly on average and typically (much) smaller.
Approaches for confidence intervals
While profiling of the likelihood is a common approach for generating confidence intervals for the negative binomial branching process model, we found that univariate profiling produced CIs with poor coverage properties for much of parameter space. Except near , the parametric bootstrap (repeatedly simulating data from the fit model and fitting to the simulated data) provides notably better coverage. Thus the default method for CI generation is a hybrid approach, using the parametric bootstrap unless the profile-based interval would be superior. This is implemented by checking whether the profiling confidence interval crosses 1, in which case it is used, or not, in which case parametric bootstrapping is used.