MIRA: Portable, Interactive Application for High-Quality Influenza, SARS-CoV-2 and RSV Genome Assembly, Annotation, and Curation
General disclaimer This repository was created for use by CDC programs to collaborate on public health related projects in support of the CDC mission. GitHub is not hosted by the CDC, but is a third party website used by CDC and its partners to share information and collaborate on software. CDC use of GitHub does not imply an endorsement of any one particular service, product, or enterprise.
Overview
MIRA is a bioinformatics pipeline that assembles Influenza genomes, SARS-CoV-2 genomes, the SARS-CoV-2 spike-gene and RSV genomes when given the raw fastq files and a samplesheet. MIRA can assemble reads from both Illumina and OxFord Nanopore sequencing machines. Coming soon, it will automate sequence submission to NCBI’s Genbank, BioSample, and SRA, as well as GISAID using SeqSender.
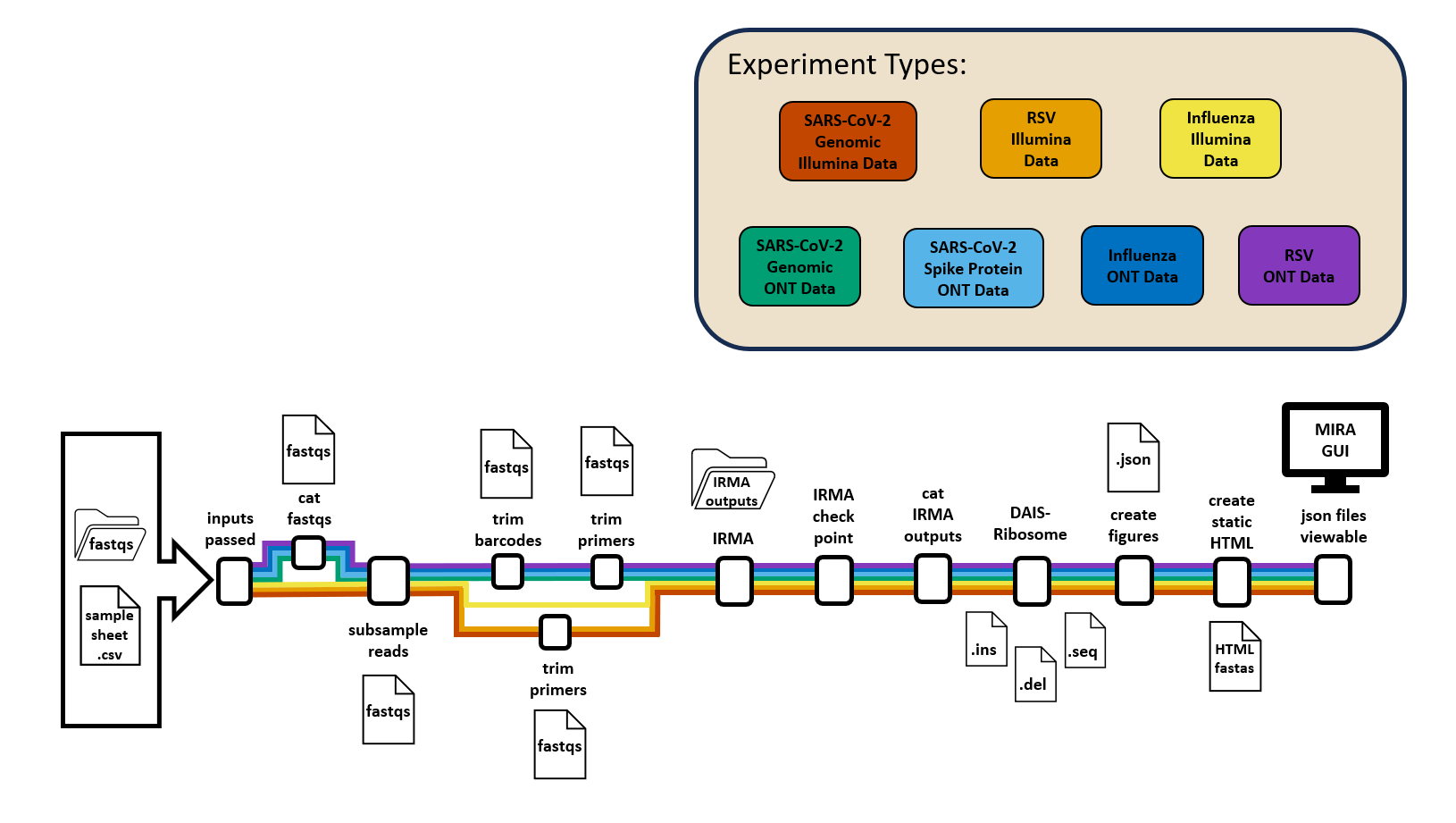
Which MIRA is right for you?
All versions of MIRA process raw sequencing reads into consensus genomes using the same pipeline and quality control. All versions create an interactive quality report and multi-fastas ready for public database submission.
Running MIRA with Docker Desktop (MIRA-GUI)
MIRA-GUI is the most popular choice for laboratory scientists. It runs on a local computer and is ideal for users who are not comfortable using the command line.
Computational Requirements:
- A minimum of 16GB of memory is required. >=32GB is recommended.
- A minimum of 8 CPU cores. 16 are recommended.
Skill Needed:
- Ability to install software on a computer.
Key features:
- No command line needed!
- Easy to use Graphical User Interface (GUI).
To run MIRA with Docker Desktop you can get started here.
Running MIRA on the command line (MIRA-CLI) with Docker
MIRA-CLI runs on a local computer from the command line. It is ideal for users who are familiar with running scripts from a terminal and are interested in automating MIRA analyses into local pipelines.
Computational Requirements:
- A minimum of 16GB of memory is required. >=32GB is recommended.
- A minimum of 8 CPU cores. 16 are recommended.
- Administrative privileges are required on a Windows operating system to run linux.
- A linux/unix (includes MacOS*) operating system is required.
- This software does work on Apple’s M-chip based OS, however we have observed intermittent instability issues.
- If you are using a Mac, go to Mac Getting Started.
Skill Needed:
- Ability to navigate your command line and run commands.
Key features:
- Interactive and sharable HTML reports.
To run MIRA-CLI you can get started with our Quick Start for Ubuntu instructions, PC instructions or Mac instructions.
Running MIRA-NF with Nextflow
MIRA-NF runs on a high performing computing (HPC) cluster or in a cloud computing platform. It is ideal for users who have access to an HPC and want to process a large volume of samples.
Computational Requirements:
- Access to an HPC or cloud computing platform.
Skill Needed:
- Familiarity with analyzing data in HPC and cloud computing platforms.
- Strong understanding of command line interface.
Key features:
- Able to run large data sets without the need for subsampling.
- Interactive and sharable HTML reports.
To run MIRA-NF you can get started by cloning the MIRA-NF Repo!
Test your MIRA Setup
- Click here to download tiny test data from ONT Influenza genome and SARS-CoV-2-spike - 40Mb
- Click here for the above data set + full genomes of Influenza and SARS-CoV-2 from Illumina MiSeqs - 1Gb
- unzip the file and find two folders:
tiny_test_run_flutiny_test_run_sc2
- move these folders into
MIRA_NGS- if you cannot find the MIRA_NGS folder in your Linux section of file explorer, look in
Linux –> home –> your username
- if you cannot find the MIRA_NGS folder in your Linux section of file explorer, look in