Install Docker Desktop
Docker allows you to run software inside an isolated “container image” on your computer with all of that application’s needed dependencies. Make sure to install the version for your operating system.
MIRA Installation
- Open Docker Desktop, and once it is running:
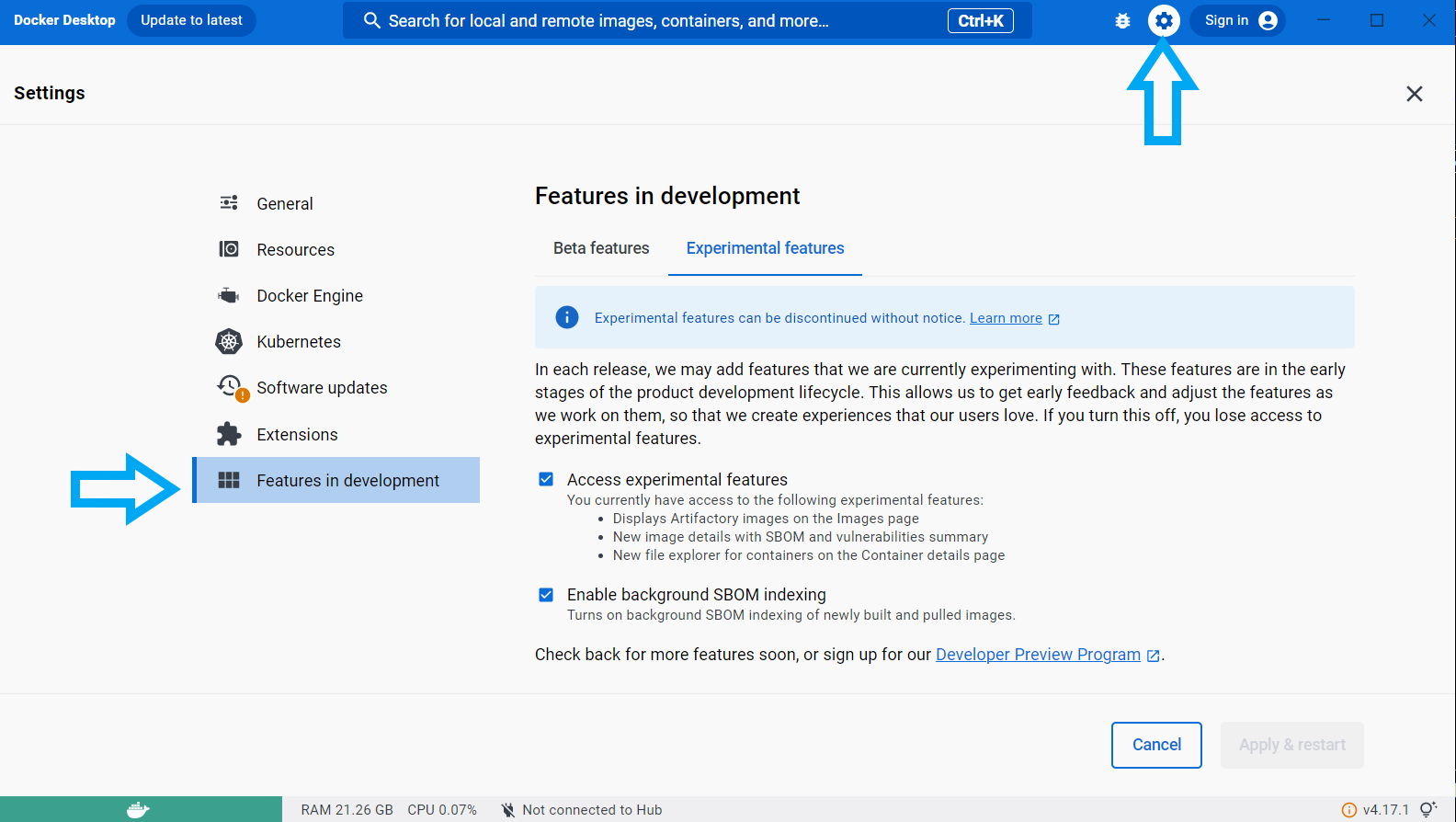
Open Settings -> Features in development -> Experimental Features:
- Ensure “Access Experimental Features” and “Enable backround SBOM Indexing” are both checked on
Next, in Beta Features, enable “Use Rosetta for x86/amd66 emulation on Apple Silicon”. This will help MIRA run faster and remain more stable
Save these settings by clicking “Apply & restart”
Open the terminal, and copy/paste these commands
*NOTE: This MIRA_NGS folder was previously called
FLU_SC2_SEQUENCING in documentation predating September
2024
curl https://raw.githubusercontent.com/CDCgov/MIRA/prod/docker-compose-cdcgov.yml | sed "s%/path/to/data%$(pwd)/%g" > docker-compose.ymlIf the docker compose command fails with a permissions error, please
try again with sudo added before the command.
You are now running MIRA! You can proceed to Running MIRA with Oxford Nanopore data or Illumina data
If these instructions show errors, please refer to our Troubleshooting page or email us at idseqsupport@cdc.gov
Test your MIRA Setup
- Click here to download tiny test data from ONT Influenza genome and SARS-CoV-2-spike - 40Mb
- Click here for the above data set + full genomes of Influenza and SARS-CoV-2 from Illumina MiSeqs - 1Gb
- unzip the file and find two folders:
tiny_test_run_flutiny_test_run_sc2
- move these 2b folders into
MIRA_NGSso that the file structure looks like:/MIRA_NGS/tiny_test_run_flu/fastq_pass/barcode##s/and/MIRA_NGS/tiny_test_run_sc2/fastq_pass/barcode##s/
 Source:
Source: