Fitting a basic renewal model
This tutorial shows the steps to build a simple renewal model featuring a latent infection process, a random walk \(\mathcal{R}(t)\) process, and an observation process for the reported infections.
We start by loading the needed components to build a basic renewal model:
import jax.numpy as jnp
import numpy as np
import numpyro
import numpyro.distributions as dist
from pyrenew.process import RandomWalk
from pyrenew.latent import (
Infections,
InfectionInitializationProcess,
InitializeInfectionsZeroPad,
)
from pyrenew.observation import PoissonObservation
from pyrenew.deterministic import DeterministicPMF
from pyrenew.model import RtInfectionsRenewalModel
from pyrenew.metaclass import RandomVariable
from pyrenew.randomvariable import DistributionalVariable, TransformedVariable
import pyrenew.transformation as t
from numpyro.infer.reparam import LocScaleReparam
By default, XLA (which is used by JAX for compilation) considers all CPU cores as one device. Depending on your system’s configuration, we recommend using numpyro’s set_host_device_count() function to set the number of devices available for parallel computing. Here, we set the device count to be 2.
numpyro.set_host_device_count(2)
pyrenew core metaclasses
Most objects in pyrenew can be treated as random variables we can
sample. At the top-level pyrenew has two metaclasses from which most
objects inherit: RandomVariable and Model. These, in turn, provide
the following sub-modules.
deterministic-RandomVariableclasses for fixed transformations and non-random parameterslatent-RandomVariableclasses for hidden states requiring statistical inferenceobservation-RandomVariableclasses for observed data and likelihoodsprocess-RandomVariableclasses for stochastic processes for disease dynamicsmodels-Modelclasses for specific epidemiological models and workflows
Architecture of RtInfectionsRenewalModel
The class RtInfectionsRenewalModel is basic renewal model consisting
of infections and reproduction numbers. It models the real-time
reproductive number \(\mathcal{R}(t)\), the average number of secondary
infections caused by an infected individual, as a renewal process model
defined in terms of:
-
generation interval, the times between infections
-
initial infections, occurring prior to time \(t = 0\)
-
\(\mathcal{R}(t)\), the time-varying reproductive number,
-
latent infections, i.e., those infections which are known to exist but are not observed (or not observable), and
-
observed infections, a subset of underlying true infections that are reported, perhaps via hospital admissions, physician’s office visits, or routine biosurveillance.
The following diagram shows a detailed view of how metaclasses, modules,
and classes are used to in the RtInfectionsRenewalModel.
flowchart LR
rand("(RandomVariable metaclass)")
models("(Model metaclass)")
subgraph observations["Observations module"]
obs["infection_obs_process_rv <br/> (PoissonObservation)"]
end
subgraph latent["Latent module"]
inf["latent_infections_rv <br/> (Infections)"]
i0["I0_rv <br/> (DistributionalVariable)"]
end
subgraph process["Process module"]
rt["Rt_process_rv <br/> (Custom class built using RandomWalk)"]
end
subgraph deterministic["Deterministic module"]
detpmf["gen_int_rv <br/> (DeterministicPMF)"]
end
subgraph model[Model module]
model1["model1 <br/> (RtInfectionsRenewalModel)"]
end
rand-->|Inherited by|observations
rand-->|Inherited by|process
rand-->|Inherited by|latent
rand-->|Inherited by|deterministic
models-->|Inherited by|model
detpmf-->|Composes|model1
i0-->|Composes|model1
rt-->|Composes|model1
obs-->|Composes|model1
inf-->|Composes|model1Implementing a RtInfectionsRenewalModel
In this example we specify a renewal model with the following components.
-
The generation interval is provided as a deterministic instance of
RandomVariable -
The
InfectionInitializationProcessspecifies the number of latent infections immediately before the renewal process begins as following a log-normal distribution with mean = 0 and standard deviation = 1. By specifyingInitializeInfectionsZeroPad, the latent infections before this time are assumed to be 0. -
A process to represent \(\mathcal{R}(t)\) as a random walk on the log scale, with an inferred initial value and a fixed Normal step-size distribution. For this, we construct a custom
RandomVariable,RtRandomWalk. -
An instance of the
Infectionsclass with default values. -
An instance of the
PoissonObservationclass with default values
# (1) The generation interval (deterministic)
pmf_array = jnp.array([0.4, 0.3, 0.2, 0.1])
gen_int = DeterministicPMF(name="gen_int", value=pmf_array)
# (2) Initial infections (inferred with a prior)
I0 = InfectionInitializationProcess(
"I0_initialization",
DistributionalVariable(name="I0", distribution=dist.LogNormal(2.5, 1)),
InitializeInfectionsZeroPad(pmf_array.size),
)
# (3) The random walk process.
# We construct a custom RandomVariable on log Rt, with an inferred s.d.
class RtRandomWalk(RandomVariable):
def validate(self):
pass
def sample(self, n: int, **kwargs) -> tuple:
sd_rt = numpyro.sample("Rt_random_walk_sd", dist.HalfNormal(0.025))
rt_rv = TransformedVariable(
name="log_rt_random_walk",
base_rv=RandomWalk(
name="log_rt",
step_rv=DistributionalVariable(
name="rw_step_rv", distribution=dist.Normal(0, sd_rt)
),
),
transforms=t.ExpTransform(),
)
rt_init_rv = DistributionalVariable(
name="init_log_rt", distribution=dist.Normal(0, 0.2)
)
init_rt = rt_init_rv.sample()
return rt_rv.sample(n=n, init_vals=init_rt, **kwargs)
rt_proc = RtRandomWalk()
# (4) Latent infection process (which will use 1 and 2)
latent_infections = Infections()
# (5) The observed infections process (with mean at the latent infections)
observation_process = PoissonObservation("poisson_rv")
With these five pieces, we can build the basic renewal model as an
instance of the RtInfectionsRenewalModel class:
model1 = RtInfectionsRenewalModel(
gen_int_rv=gen_int,
I0_rv=I0,
Rt_process_rv=rt_proc,
latent_infections_rv=latent_infections,
infection_obs_process_rv=observation_process,
)
Using numpyro, we can simulate data using the sample() member
function of RtInfectionsRenewalModel. The sample() method of the
RtInfectionsRenewalModel class returns a list composed of the Rt and
infections sequences, called sim_data:
with numpyro.handlers.seed(rng_seed=53):
sim_data = model1.sample(n_datapoints=40)
sim_data
RtInfectionsRenewalSample(Rt=[1.0288756 1.0117078 1.0572513 1.0822552 1.0788958 1.0905282 1.0858724
1.0702966 1.048627 1.0500144 1.0381658 1.016077 1.0067611 1.0017354
0.9862044 0.9754583 1.0032375 1.0138954 1.016938 1.0482056 1.0621839
1.0489949 1.0511138 1.0635226 1.07795 1.0862241 1.1069213 1.1032605
1.1220009 1.0832963 1.099409 1.0937598 1.1113575 1.0927193 1.0821252
1.0777551 1.0788382 1.0901768 1.0664136 1.0852975], latent_infections=[0. 0. 0. 4.286751 1.7642133 2.0150292 2.3181567
2.5035694 2.4558926 2.6156998 2.731596 2.8029823 2.841151 2.9245467
2.9649189 2.9586742 2.9618607 2.9629183 2.9210904 2.8732626 2.9238374
2.9524875 2.9744496 3.0897117 3.1983426 3.2480965 3.3363705 3.4645493
3.617799 3.7785044 4.0106955 4.201044 4.489246 4.5888443 4.8633347
5.0749807 5.396403 5.5866446 5.794682 6.0145683 6.2585573 6.568745
6.705019 7.0607276], observed_infections=[ 1 0 0 2 1 2 1 2 1 3 1 5 6 2 5 1 3 3 3 1 2 2 2 4
2 2 4 3 7 4 6 3 6 4 5 5 2 6 14 7])
To understand what has been accomplished here, visualize an \(\mathcal{R}(t)\) sample path (left panel) and infections over time (right panel):
import matplotlib.pyplot as plt
fig, axs = plt.subplots(1, 2)
# Rt plot
axs[0].plot(sim_data.Rt)
axs[0].set_ylabel("Rt")
# Infections plot
axs[1].plot(sim_data.observed_infections)
axs[1].set_ylabel("Infections")
fig.suptitle("Basic renewal model")
fig.supxlabel("Time")
plt.tight_layout()
plt.show()
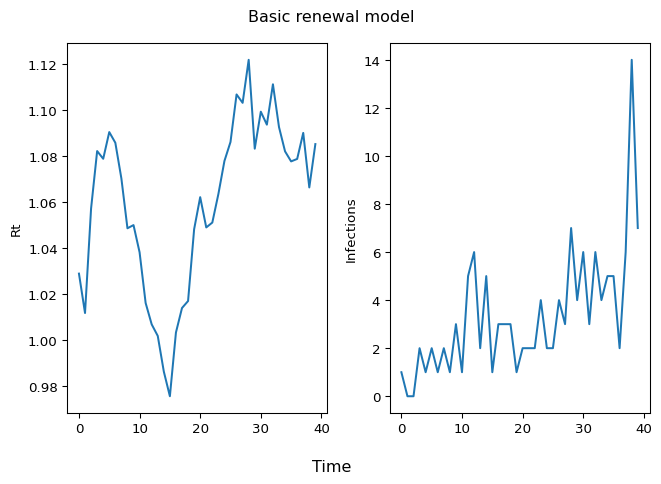
Figure 1: Rt and Infections
To fit the model, we can use the run() method of the
RtInfectionsRenewalModel class (an inherited method from the metaclass
Model). model1.run() will call the run method of the model1
object, which will generate an instance of model MCMC simulation, with
2000 warm-up iterations for the MCMC algorithm, used to tune the
parameters of the MCMC algorithm to improve efficiency of the sampling
process. From the posterior distribution of the model parameters, 1000
samples will be drawn and used to estimate the posterior distributions
and compute summary statistics. Observed data is provided to the model
using the sim_data object previously generated. mcmc_args provides
additional arguments for the MCMC algorithm.
import jax
model1.run(
num_warmup=2000,
num_samples=1000,
data_observed_infections=sim_data.observed_infections,
rng_key=jax.random.key(54),
mcmc_args=dict(progress_bar=False, num_chains=2),
)
Now, let’s investigate the output, particularly the posterior distribution of the \(\mathcal{R}(t)\) estimates:
import arviz as az
# Create arviz inference data object
idata = az.from_numpyro(
posterior=model1.mcmc,
)
# Extract Rt signal samples across chains
rt = az.extract(idata.posterior["Rt"], num_samples=100)["Rt"].values
# Plot Rt signal
fig, ax = plt.subplots(1, 1, figsize=(8, 6))
ax.plot(
np.arange(rt.shape[0]),
rt,
color="skyblue",
alpha=0.10,
)
ax.plot([], [], color="skyblue", alpha=0.05, label="Rt Posterior Samples")
ax.plot(
np.arange(rt.shape[0]),
rt.mean(axis=1),
color="black",
linewidth=2.0,
linestyle="--",
label="Sample Mean",
)
ax.legend(loc="best")
ax.set_ylabel(r"$\mathscr{R}_t$ Signal", fontsize=20)
ax.set_xlabel("Days", fontsize=20)
plt.show()
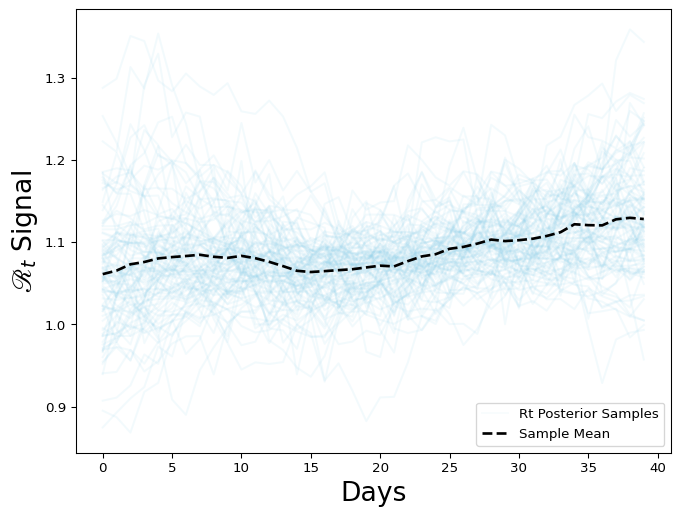
Figure 2: Rt posterior distribution
We can use the get_samples method to extract samples from the model
Rt_samp = model1.mcmc.get_samples()["Rt"]
latent_infection_samp = model1.mcmc.get_samples()["all_latent_infections"]
We can also convert the fitted model to ArviZ InferenceData object and use ArviZ package to extarct samples, calculate statistics, create model diagnostics and visualizations.
import arviz as az
idata = az.from_numpyro(model1.mcmc)
and use the InferenceData to compute the model-fit diagnostics. Here, we show diagnostic summary for the first 10 effective reproduction number \(\mathcal{R}(t)\).
diagnostic_stats_summary = az.summary(
idata.posterior["Rt"][::, ::, 4:], # ignore nan padding
kind="diagnostics",
)
print(diagnostic_stats_summary)
mcse_mean mcse_sd ess_bulk ess_tail r_hat
Rt[4] 0.002 0.003 1427.0 950.0 1.01
Rt[5] 0.002 0.003 1385.0 784.0 1.02
Rt[6] 0.002 0.003 1292.0 744.0 1.01
Rt[7] 0.002 0.003 1312.0 761.0 1.02
Rt[8] 0.002 0.003 1097.0 579.0 1.01
Rt[9] 0.002 0.003 1145.0 652.0 1.01
Rt[10] 0.001 0.002 1280.0 775.0 1.01
Rt[11] 0.001 0.002 1647.0 939.0 1.01
Rt[12] 0.001 0.002 1894.0 891.0 1.01
Rt[13] 0.001 0.002 1604.0 633.0 1.01
Rt[14] 0.001 0.003 1173.0 692.0 1.01
Rt[15] 0.002 0.002 625.0 469.0 1.01
Rt[16] 0.002 0.002 566.0 694.0 1.01
Rt[17] 0.002 0.002 398.0 592.0 1.01
Rt[18] 0.003 0.002 295.0 542.0 1.01
Rt[19] 0.002 0.002 328.0 472.0 1.01
Rt[20] 0.002 0.002 448.0 598.0 1.01
Rt[21] 0.002 0.003 607.0 511.0 1.01
Rt[22] 0.001 0.003 911.0 482.0 1.02
Rt[23] 0.001 0.002 1564.0 568.0 1.01
Rt[24] 0.001 0.002 2295.0 863.0 1.01
Rt[25] 0.001 0.002 2789.0 685.0 1.01
Rt[26] 0.001 0.002 3396.0 695.0 1.01
Rt[27] 0.001 0.002 3006.0 698.0 1.01
Rt[28] 0.001 0.002 2551.0 869.0 1.01
Rt[29] 0.001 0.002 1783.0 878.0 1.00
Rt[30] 0.001 0.002 2108.0 825.0 1.00
Rt[31] 0.001 0.002 1586.0 1230.0 1.00
Rt[32] 0.001 0.002 1275.0 657.0 1.01
Rt[33] 0.001 0.002 1036.0 504.0 1.00
Rt[34] 0.002 0.002 821.0 984.0 1.01
Rt[35] 0.002 0.002 668.0 636.0 1.00
Rt[36] 0.002 0.002 669.0 775.0 1.00
Rt[37] 0.003 0.003 471.0 709.0 1.01
Rt[38] 0.004 0.003 357.0 410.0 1.00
Rt[39] 0.004 0.004 468.0 663.0 1.01
Below we use plot_trace to inspect the trace of the first 10 inferred
\(\mathcal{R}(t)\) values.
plt.rcParams["figure.constrained_layout.use"] = True
az.plot_trace(
idata.posterior,
var_names=["Rt"],
coords={"Rt_dim_0": np.arange(4, 14)},
compact=False,
)
plt.show()
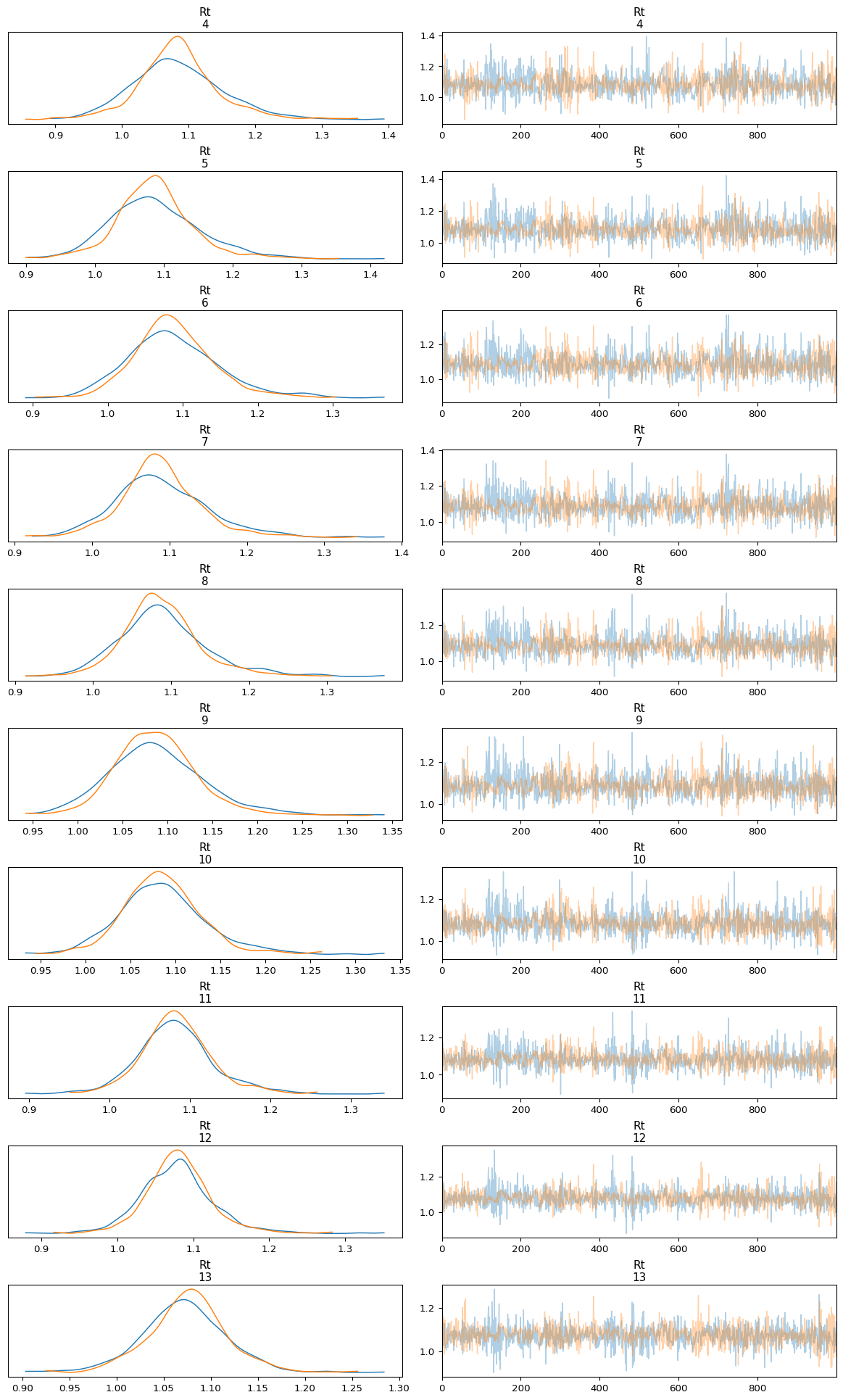
Figure 3: Trace plot of Rt posterior distribution
We inspect the posterior distribution of \(\mathcal{R}(t)\) by plotting the 90% and 50% highest density intervals:
x_data = idata.posterior["Rt_dim_0"][4:]
y_data = idata.posterior["Rt"][::, ::, 4:]
fig, axes = plt.subplots(figsize=(6, 5))
az.plot_hdi(
x_data,
y_data,
hdi_prob=0.9,
color="C0",
fill_kwargs={"alpha": 0.3},
ax=axes,
)
az.plot_hdi(
x_data,
y_data,
hdi_prob=0.5,
color="C0",
fill_kwargs={"alpha": 0.6},
ax=axes,
)
# Add mean of the posterior to the figure
median_ts = y_data.median(dim=["chain", "draw"])
axes.plot(x_data, median_ts, color="C0", label="Median")
axes.legend()
axes.set_title("Posterior Effective Reproduction Number", fontsize=10)
axes.set_xlabel("Time", fontsize=10)
axes.set_ylabel("$\\mathcal{R}(t)$", fontsize=10)
plt.show()
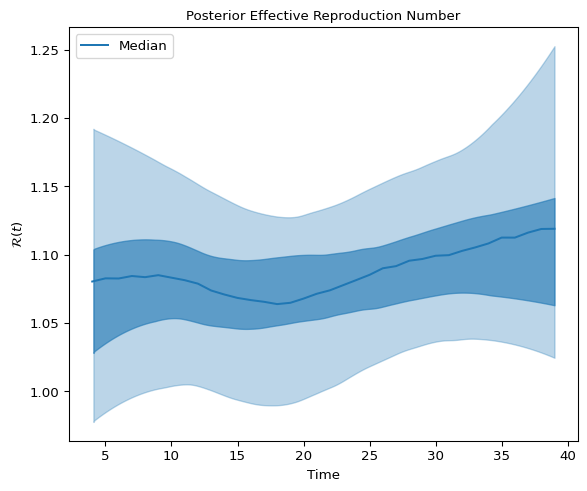
Figure 4: High density interval for Effective Reproduction Number
and latent infections:
x_data = idata.posterior["all_latent_infections_dim_0"]
y_data = idata.posterior["all_latent_infections"]
fig, axes = plt.subplots(figsize=(6, 5))
az.plot_hdi(
x_data,
y_data,
hdi_prob=0.9,
color="C0",
smooth=False,
fill_kwargs={"alpha": 0.3},
ax=axes,
)
az.plot_hdi(
x_data,
y_data,
hdi_prob=0.5,
color="C0",
smooth=False,
fill_kwargs={"alpha": 0.6},
ax=axes,
)
# plot the posterior median
median_ts = y_data.median(dim=["chain", "draw"])
axes.plot(x_data, median_ts, color="C0", label="Median")
axes.legend()
axes.set_title("Posterior Latent Infections", fontsize=10)
axes.set_xlabel("Time", fontsize=10)
axes.set_ylabel("Latent Infections", fontsize=10)
plt.show()
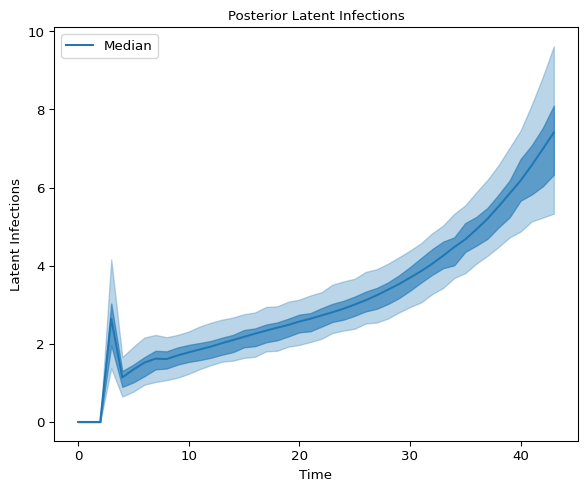
Figure 5: High density interval for Latent Infections