Getting started with pyrenew#
pyrenew is a flexible tool for simulating and making statistical
inferences from epidemiologic models, with an emphasis on renewal
models. Built on numpyro, pyrenew provides core components for
model building and pre-defined models for processing various
observational processes. This document illustrates how pyrenew can
be used to build a basic renewal model.
The fundamentals#
pyrenew’s core components are the metaclasses RandomVariable
and Model (in Python, a metaclass is a class whose instances are
also classes, where a class is a template for making objects). Within
the pyrenew package, a RandomVariable is a quantity that models
can estimate and sample from, including deterministic quantities.
The benefit of this design is that the definition of the sample()
function can be arbitrary, allowing the user to either sample from a
distribution using numpyro.sample(), compute fixed quantities (like
a mechanistic equation), or return a fixed value (like a pre-computed
PMF.) For instance, when estimating a PMF, the RandomVariable
sampling function may roughly be defined as:
# define a new class called MyRandVar that inherits from the RandomVariable class
class MyRandVar(RandomVariable):
#define a method called sample that returns an object of type ArrayLike
def sample(...) -> ArrayLike:
# calls sample function from NumPyro package
return numpyro.sample(...)
Whereas, in some other cases, we may instead use a fixed quantity for
that variable (like a pre-computed PMF), where the
RandomVariable’s sample function could instead be defined as:
# instead define MyRandVar to still inherit from the RandVariable class
class MyRandVar(RandomVariable):
#define sample method that still returns an ArrayLike object
def sample(...) -> ArrayLike:
#sampling method is a pre-computed PMF, a JAX NumPy array with explicit elements
return jax.numpy.array([0.2, 0.7, 0.1])
Thus, when a Model samples from MyRandVar, it could be either
adding random variables to be estimated (first case) or just retrieving
some quantity needed for other calculations (second case.)
The Model metaclass provides basic functionality for estimating and
simulation. Like RandomVariable, the Model metaclass has a
sample() method that defines the model structure. Ultimately, models
can be nested (or inherited), providing a straightforward way to add
layers of complexity.
‘Hello world’ model#
This section will show the steps to build a simple renewal model featuring a latent infection process, a random walk Rt process, and an observation process for the reported infections.
We start by loading the needed components to build a basic renewal model:
import jax.numpy as jnp
import numpy as np
import numpyro as npro
import numpyro.distributions as dist
from pyrenew.process import RtRandomWalkProcess
from pyrenew.latent import (
Infections,
InfectionSeedingProcess,
SeedInfectionsZeroPad,
)
from pyrenew.observation import PoissonObservation
from pyrenew.deterministic import DeterministicPMF
from pyrenew.model import RtInfectionsRenewalModel
from pyrenew.metaclass import DistributionalRV
import pyrenew.transformation as t
The pyrenew package models the real-time reproductive number \(R_t\), the ratio of new infections at time \(t\) to previous infections at some time \(t-s\), as a renewal process model. Our basic renewal process model defines five components:
generation interval, the times between infections
initial infections, occurring prior to time \(t = 0\)
\(R_t\), the real-time reproductive number,
latent infections, i.e., those infections which are known to exist but are not observed (or not observable), and
observed infections, a subset of underlying true infections that are reported, perhaps via hospital admissions, physician’s office visits, or routine biosurveillance.
To initialize these five components within the renewal modeling framework, we estimate each component with:
In this example, the generation interval is not estimated but passed as a deterministic instance of
RandomVariablean instance of the
InfectionSeedingProcessclass, where the number of latent infections immediately before the renewal process begins follows a log-normal distribution with mean = 0 and standard deviation = 1. By specifyingSeedInfectionsZeroPad, the latent infections before this time are assumed to be 0.an instance of the
RtRandomWalkProcessclass with default valuesan instance of the
Infectionsclass with default values, andan instance of the
PoissonObservationclass with default values
# (1) The generation interval (deterministic)
pmf_array = jnp.array([0.25, 0.25, 0.25, 0.25])
gen_int = DeterministicPMF(pmf_array, name="gen_int")
# (2) Initial infections (inferred with a prior)
I0 = InfectionSeedingProcess(
"I0_seeding",
DistributionalRV(dist=dist.LogNormal(0, 1), name="I0"),
SeedInfectionsZeroPad(pmf_array.size),
)
# (3) The random process for Rt
rt_proc = RtRandomWalkProcess(
Rt0_dist=dist.TruncatedNormal(loc=1.2, scale=0.2, low=0),
Rt_transform=t.ExpTransform().inv,
Rt_rw_dist=dist.Normal(0, 0.025),
)
# (4) Latent infection process (which will use 1 and 2)
latent_infections = Infections()
# (5) The observed infections process (with mean at the latent infections)
observation_process = PoissonObservation()
With these five pieces, we can build the basic renewal model as an
instance of the RtInfectionsRenewalModel class:
model1 = RtInfectionsRenewalModel(
gen_int_rv=gen_int,
I0_rv=I0,
Rt_process_rv=rt_proc,
latent_infections_rv=latent_infections,
infection_obs_process_rv=observation_process,
)
The following diagram summarizes how the modules interact via
composition; notably, gen_int, I0, rt_proc,
latent_infections, and observed_infections are instances of
RandomVariable, which means these can be easily replaced to generate
a different instance of the RtInfectionsRenewalModel class:
Using numpyro, we can simulate data using the sample() member
function of RtInfectionsRenewalModel. The sample() method of the
RtInfectionsRenewalModel class returns a list composed of the Rt
and infections sequences, called sim_data:
np.random.seed(223)
with npro.handlers.seed(rng_seed=np.random.randint(1, 60)):
sim_data = model1.sample(n_timepoints_to_simulate=30)
sim_data
RtInfectionsRenewalSample(Rt=[ nan nan nan nan 1.2022278 1.2111099 1.2325984
1.2104921 1.2023039 1.1970979 1.2384264 1.2423582 1.245498 1.241344
1.2081108 1.1938375 1.2711959 1.3189521 1.3054799 1.3317624 1.3068875
1.3177233 1.2765354 1.3001204 1.3273207 1.3038 1.2788019 1.2726372
1.2690852 1.3244348 1.263264 1.2540829 1.2211404 1.247711 ], latent_infections=[ 0. 0. 0. 7.7240214 2.3215084 3.0415602
4.0327816 5.180868 4.381411 4.978916 5.750626 6.3024273
6.66758 7.354823 7.8755097 8.416656 9.633939 10.973987
12.043082 13.673094 15.135098 17.072838 18.485544 20.921074
23.76387 26.155312 28.5575 31.624323 34.93189 40.15323
42.71946 46.849056 50.266296 56.14326 ], observed_infections=[ 0 0 0 0 2 3 5 5 1 3 4 7 6 8 7 7 18 9 10 11 16 19 14 24
17 34 34 25 38 39 43 52 50 51])
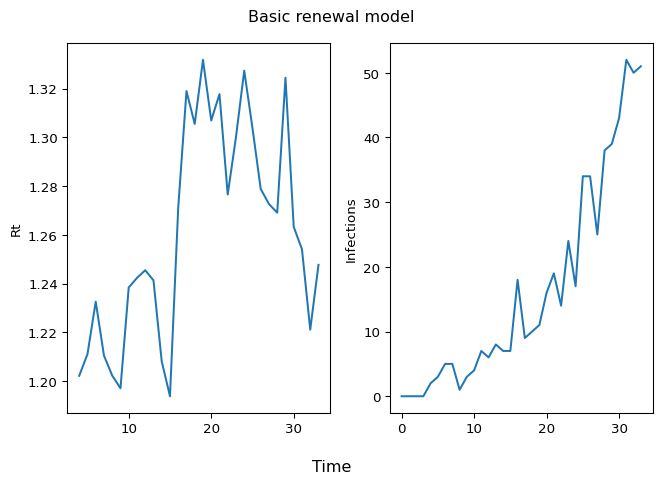
To understand what has been accomplished here, visualize an \(R_t\) sample path (left panel) and infections over time (right panel):
import matplotlib.pyplot as plt
fig, axs = plt.subplots(1, 2)
# Rt plot
axs[0].plot(sim_data.Rt)
axs[0].set_ylabel("Rt")
# Infections plot
axs[1].plot(sim_data.observed_infections)
axs[1].set_ylabel("Infections")
fig.suptitle("Basic renewal model")
fig.supxlabel("Time")
plt.tight_layout()
plt.show()
To fit the model, we can use the run() method of the
RtInfectionsRenewalModel class (an inherited method from the
metaclass Model). model1.run() will call the run method of
the model1 object, which will generate an instance of model MCMC
simulation, with 2000 warm-up iterations for the MCMC algorithm, used to
tune the parameters of the MCMC algorithm to improve efficiency of the
sampling process. From the posterior distribution of the model
parameters, 1000 samples will be drawn and used to estimate the
posterior distributions and compute summary statistics. Observed data is
provided to the model using the sim_data object previously
generated. mcmc_args provides additional arguments for the MCMC
algorithm.
import jax
model1.run(
num_warmup=2000,
num_samples=1000,
data_observed_infections=sim_data.observed_infections,
rng_key=jax.random.PRNGKey(54),
mcmc_args=dict(progress_bar=False, num_chains=2),
)
/home/runner/work/multisignal-epi-inference/multisignal-epi-inference/model/src/pyrenew/metaclass.py:304: UserWarning: There are not enough devices to run parallel chains: expected 2 but got 1. Chains will be drawn sequentially. If you are running MCMC in CPU, consider using `numpyro.set_host_device_count(2)` at the beginning of your program. You can double-check how many devices are available in your system using `jax.local_device_count()`.
self.mcmc = MCMC(
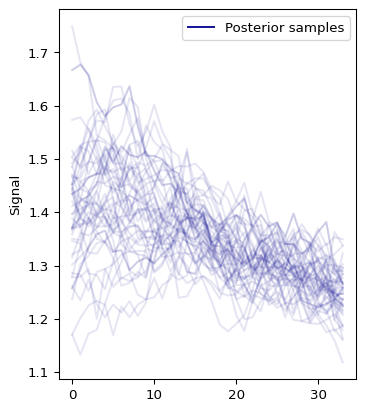
Now, let’s investigate the output, particularly the posterior distribution of the \(R_t\) estimates:
out = model1.plot_posterior(var="Rt")
We can use ArviZ package to create model diagnostics and visualizations. We start by converting the fitted model to ArviZ InferenceData object:
import arviz as az
idata = az.from_numpyro(model1.mcmc)
and use the InferenceData to compute the model-fit diagnostics. Here, we show diagnostic summary for the first 10 effective reproduction number \(R_t\).
diagnostic_stats_summary = az.summary(
idata.posterior["Rt"],
kind="diagnostics",
)
print(diagnostic_stats_summary[:10])
mcse_mean mcse_sd ess_bulk ess_tail r_hat
Rt[0] 0.003 0.002 1309.0 1339.0 1.0
Rt[1] 0.003 0.002 1331.0 1264.0 1.0
Rt[2] 0.003 0.002 1340.0 1395.0 1.0
Rt[3] 0.003 0.002 1421.0 1335.0 1.0
Rt[4] 0.002 0.002 1459.0 1464.0 1.0
Rt[5] 0.002 0.002 1597.0 1303.0 1.0
Rt[6] 0.002 0.002 1539.0 1454.0 1.0
Rt[7] 0.002 0.001 1707.0 1529.0 1.0
Rt[8] 0.002 0.001 1494.0 1674.0 1.0
Rt[9] 0.002 0.001 1951.0 1517.0 1.0
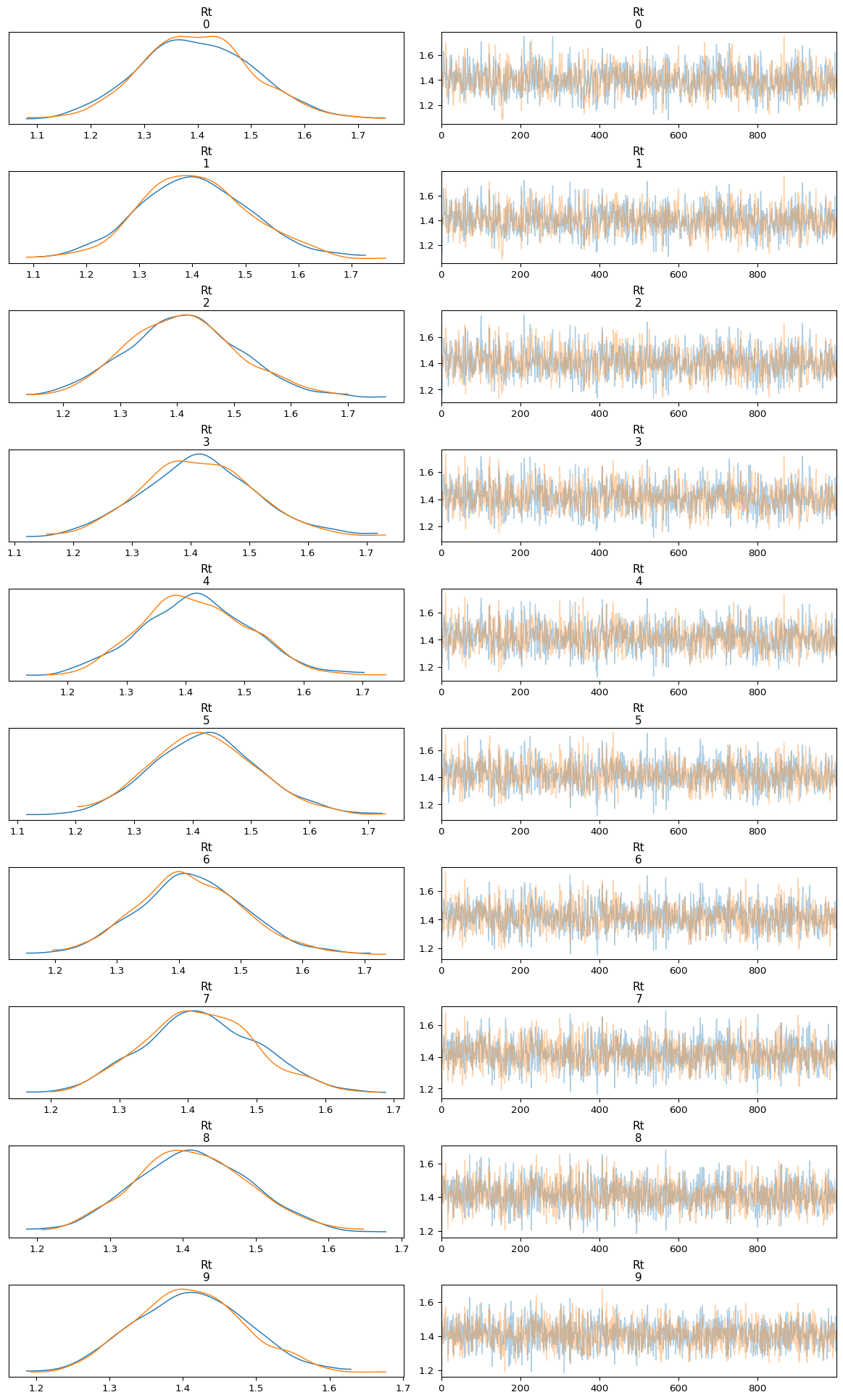
Below we use plot_trace to inspect the trace of the first 10
\(R_t\) estimates.
plt.rcParams["figure.constrained_layout.use"] = True
az.plot_trace(
idata.posterior,
var_names=["Rt"],
coords={"Rt_dim_0": np.arange(10)},
compact=False,
);
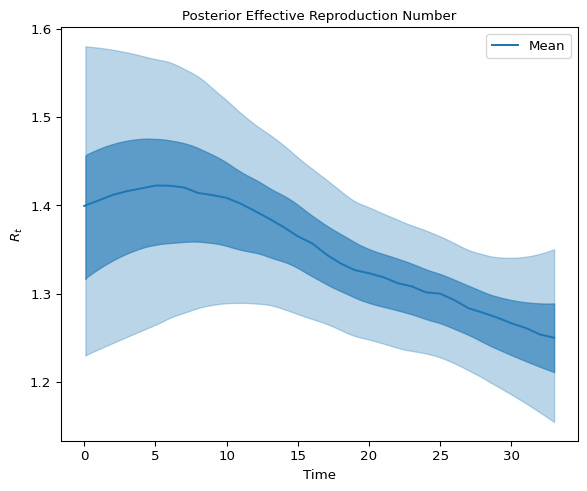
We inspect the posterior distribution of \(R_t\) by plotting the 90% and 50% highest density intervals:
x_data = idata.posterior["Rt_dim_0"]
y_data = idata.posterior["Rt"]
fig, axes = plt.subplots(figsize=(6, 5))
az.plot_hdi(
x_data,
y_data,
hdi_prob=0.9,
color="C0",
fill_kwargs={"alpha": 0.3},
ax=axes,
)
az.plot_hdi(
x_data,
y_data,
hdi_prob=0.5,
color="C0",
fill_kwargs={"alpha": 0.6},
ax=axes,
)
# Add mean of the posterior to the figure
mean_Rt = np.mean(idata.posterior["Rt"], axis=1)
axes.plot(x_data, mean_Rt[0], color="C0", label="Mean")
axes.legend()
axes.set_title("Posterior Effective Reproduction Number", fontsize=10)
axes.set_xlabel("Time", fontsize=10)
axes.set_ylabel("$R_t$", fontsize=10);
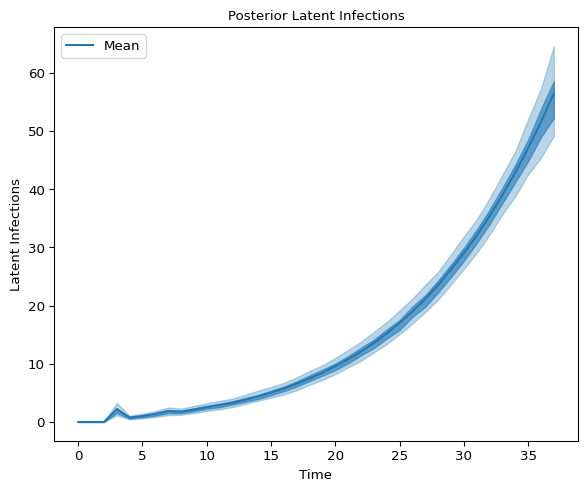
and latent infections:
x_data = idata.posterior["all_latent_infections_dim_0"]
y_data = idata.posterior["all_latent_infections"]
fig, axes = plt.subplots(figsize=(6, 5))
az.plot_hdi(
x_data,
y_data,
hdi_prob=0.9,
color="C0",
smooth=False,
fill_kwargs={"alpha": 0.3},
ax=axes,
)
az.plot_hdi(
x_data,
y_data,
hdi_prob=0.5,
color="C0",
smooth=False,
fill_kwargs={"alpha": 0.6},
ax=axes,
)
# Add mean of the posterior to the figure
mean_latent_infection = np.mean(
idata.posterior["all_latent_infections"], axis=1
)
axes.plot(x_data, mean_latent_infection[0], color="C0", label="Mean")
axes.legend()
axes.set_title("Posterior Latent Infections", fontsize=10)
axes.set_xlabel("Time", fontsize=10)
axes.set_ylabel("Latent Infections", fontsize=10);
Architecture of pyrenew#
pyrenew leverages numpyro’s flexibility to build models via
composition. As a principle, most objects in pyrenew can be treated
as random variables we can sample. At the top-level pyrenew has two
metaclasses from which most objects inherit: RandomVariable and
Model. From them, the following four sub-modules arise:
The
processsub-module,The
deterministicsub-module,The
observationsub-module,The
latentsub-module, andThe
modelssub-module
The first four are collections of instances of RandomVariable, and
the last is a collection of instances of Model. The following
diagram shows a detailed view of how metaclasses, modules, and classes
interact to create the RtInfectionsRenewalModel instantiated in the
previous section: