Extending pyrenew#
This tutorial illustrates how to extend pyrenew with custom
RandomVariable classes. We will use the InfectionsWithFeedback
class as an example. The InfectionsWithFeedback class is a
RandomVariable that models the number of infections at time
\(t\) as a function of the number of infections at time
\(t - \tau\) and the reproduction number at time \(t\). The
reproduction number at time \(t\) is a function of the unadjusted
reproduction number at time \(t - \tau\) and the number of
infections at time \(t - \tau\):
Where \(\mathcal{R}^u(t)\) is the unadjusted reproduction number, \(g(t)\) is the generation interval, \(\gamma(t)\) is the infection feedback strength, and \(f(t)\) is the infection feedback pmf.
The expected outcome#
Before we start, let’s simulate the model with the original
InfectionsWithFeedback class. To keep it simple, we will simulate
the model with no observation process, in other words, only with latent
infections. The following code-chunk loads the required libraries and
defines the model components:
import jax
import jax.numpy as jnp
import numpy as np
import numpyro as npro
import numpyro.distributions as dist
from pyrenew.deterministic import DeterministicPMF, DeterministicVariable
from pyrenew.latent import InfectionsWithFeedback
from pyrenew.model import RtInfectionsRenewalModel
from pyrenew.process import RtRandomWalkProcess
from pyrenew.metaclass import DistributionalRV
from pyrenew.latent import InfectionSeedingProcess, SeedInfectionsExponential
import pyrenew.transformation as t
/home/runner/.cache/pypoetry/virtualenvs/pyrenew-ay_vsbmF-py3.12/lib/python3.12/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
The following code-chunk defines the model components. Notice that for both the generation interval and the infection feedback, we use a deterministic PMF with equal probabilities:
gen_int_array = jnp.array([0.25, 0.5, 0.15, 0.1])
gen_int = DeterministicPMF(gen_int_array, name="gen_int")
feedback_strength = DeterministicVariable(0.05, name="feedback_strength")
I0 = InfectionSeedingProcess(
"I0_seeding",
DistributionalRV(dist=dist.LogNormal(0, 1), name="I0"),
SeedInfectionsExponential(
gen_int_array.size,
DeterministicVariable(0.5, name="rate"),
),
)
latent_infections = InfectionsWithFeedback(
infection_feedback_strength=feedback_strength,
infection_feedback_pmf=gen_int,
)
rt = RtRandomWalkProcess(
Rt0_dist=dist.TruncatedNormal(loc=1.2, scale=0.2, low=0),
Rt_transform=t.ExpTransform().inv,
Rt_rw_dist=dist.Normal(0, 0.025),
)
With all the components defined, we can build the model:
model0 = RtInfectionsRenewalModel(
gen_int_rv=gen_int,
I0_rv=I0,
latent_infections_rv=latent_infections,
Rt_process_rv=rt,
infection_obs_process_rv=None,
)
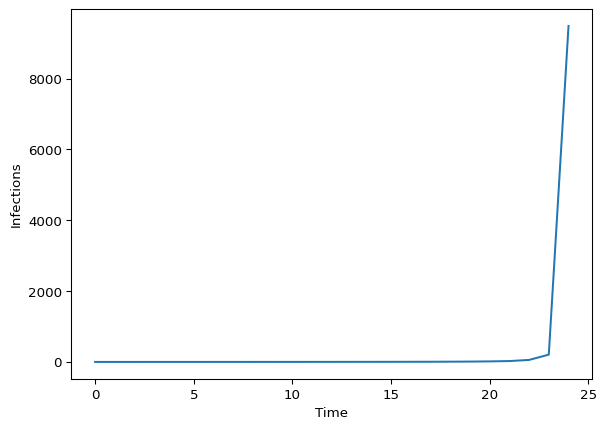
And simulate from it:
# Sampling and fitting model 0 (with no obs for infections)
np.random.seed(223)
with npro.handlers.seed(rng_seed=np.random.randint(1, 600)):
model0_samp = model0.sample(n_timepoints_to_simulate=30)
/home/runner/.cache/pypoetry/virtualenvs/pyrenew-ay_vsbmF-py3.12/lib/python3.12/site-packages/jax/_src/numpy/lax_numpy.py:3044: FutureWarning: None encountered in jnp.array(); this is currently treated as NaN. In the future this will result in an error.
return array(arys[0], copy=False, ndmin=1)
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.plot(model0_samp.latent_infections)
ax.set_xlabel("Time")
ax.set_ylabel("Infections")
plt.show()
Pyrenew’s random variable class#
Fundamentals#
All instances of PyRenew’s RandomVariable should have at least three
functions: __init__(), validate(), and sample(). The
__init__() function is the constructor and initializes the class.
The validate() function checks if the class is correctly
initialized. Finally, the sample() method contains the core of the
class; it should return a tuple or named tuple. The following is a
minimal example of a RandomVariable class based on
numpyro.distributions.Normal:
from pyrenew.metaclass import RandomVariable
class MyNormal(RandomVariable):
def __init__(self, loc, scale):
self.validate(scale)
self.loc = loc
self.scale = scale
return None
@staticmethod
def validate(self):
if self.scale <= 0:
raise ValueError("Scale must be positive")
return None
def sample(self, **kwargs):
return (dist.Normal(loc=self.loc, scale=self.scale),)
The @staticmethod decorator exposes the validate function to be
used outside the class. Next, we show how to build a more complex
RandomVariable class; the InfectionsWithFeedback class.
The InfectionsWithFeedback class#
Although returning namedtuples is not strictly required, they are the
recommended return type, as they make the code more readable. The
following code-chunk shows how to create a named tuple for the
InfectionsWithFeedback class:
from collections import namedtuple
# Creating a tuple to store the output
InfFeedbackSample = namedtuple(
typename="InfFeedbackSample",
field_names=["infections", "rt"],
defaults=(None, None),
)
The next step is to create the actual class. The bulk of its
implementation lies in the function
pyrenew.latent.compute_infections_from_rt_with_feedback(). We will
also use the pyrenew.arrayutils.pad_x_to_match_y() function to
ensure the passed vectors match their lengths. The following code-chunk
shows most of the implementation of the InfectionsWithFeedback
class:
# Creating the class
from pyrenew.metaclass import RandomVariable
from pyrenew.latent import compute_infections_from_rt_with_feedback
from pyrenew import arrayutils as au
from jax.typing import ArrayLike
import jax.numpy as jnp
class InfFeedback(RandomVariable):
"""Latent infections"""
def __init__(
self,
infection_feedback_strength: RandomVariable,
infection_feedback_pmf: RandomVariable,
) -> None:
"""Constructor"""
self.infection_feedback_strength = infection_feedback_strength
self.infection_feedback_pmf = infection_feedback_pmf
return None
def validate(self):
"""
Generally, this method should be more meaningful, but we will skip it for now
"""
return None
def sample(
self,
Rt: ArrayLike,
I0: ArrayLike,
gen_int: ArrayLike,
**kwargs,
) -> tuple:
"""Sample infections with feedback"""
# Generation interval
gen_int_rev = jnp.flip(gen_int)
# Baseline infections
I0_vec = I0[-gen_int_rev.size :]
# Sampling inf feedback strength and adjusting the shape
inf_feedback_strength, *_ = self.infection_feedback_strength.sample(
**kwargs,
)
inf_feedback_strength = au.pad_x_to_match_y(
x=inf_feedback_strength, y=Rt, fill_value=inf_feedback_strength[0]
)
# Sampling inf feedback and adjusting the shape
inf_feedback_pmf, *_ = self.infection_feedback_pmf.sample(**kwargs)
inf_fb_pmf_rev = jnp.flip(inf_feedback_pmf)
# Generating the infections with feedback
all_infections, Rt_adj = compute_infections_from_rt_with_feedback(
I0=I0_vec,
Rt_raw=Rt,
infection_feedback_strength=inf_feedback_strength,
reversed_generation_interval_pmf=gen_int_rev,
reversed_infection_feedback_pmf=inf_fb_pmf_rev,
)
# Storing adjusted Rt for future use
npro.deterministic("Rt_adjusted", Rt_adj)
# Preparing theoutput
return InfFeedbackSample(
infections=all_infections,
rt=Rt_adj,
)
The core of the class is implemented in the sample() method. Things
to highlight from the above code:
Arguments of ``sample``: The
InfFeedbackclass will be used withinRtInfectionsRenewalModelto generate latent infections. During the sampling process,InfFeedback.sample()will receive the reproduction number, the initial number of infections, and the generation interval.RandomVariable.sample()calls are expected to include the**kwargsargument, even if unused.Calls to ``RandomVariable.sample()``: All calls to
RandomVariable.sample()are expected to return a tuple or named tuple. In our implementation, we capture the output ofinfection_feedback_strength.sample()andinfection_feedback_pmf.sample()in the variablesinf_feedback_strengthandinf_feedback_pmf, respectively, disregarding the other outputs (i.e., using*_).Saving computed quantities: Since
Rt_adjis not generated vianumpyro.sample(), we usenumpyro.deterministic()to record the quantity to a site; allowing us to access it later.Return type of ``InfFeedback.sample()``: As said before, the
sample()method should return a tuple or named tuple. In our case, we return a named tupleInfFeedbackSamplewith two fields:infectionsandrt.
latent_infections2 = InfFeedback(
infection_feedback_strength=feedback_strength,
infection_feedback_pmf=gen_int,
)
model1 = RtInfectionsRenewalModel(
gen_int_rv=gen_int,
I0_rv=I0,
latent_infections_rv=latent_infections2,
Rt_process_rv=rt,
infection_obs_process_rv=None,
)
# Sampling and fitting model 0 (with no obs for infections)
np.random.seed(223)
with npro.handlers.seed(rng_seed=np.random.randint(1, 600)):
model1_samp = model1.sample(n_timepoints_to_simulate=30)
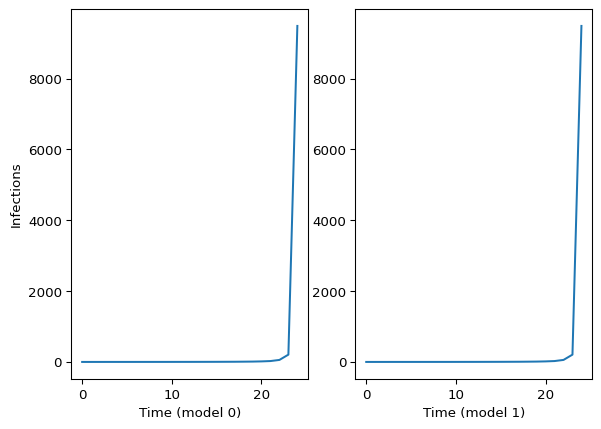
Comparing model0 with model1, these two should match:
import matplotlib.pyplot as plt
fig, ax = plt.subplots(ncols=2)
ax[0].plot(model0_samp.latent_infections)
ax[1].plot(model1_samp.latent_infections)
ax[0].set_xlabel("Time (model 0)")
ax[1].set_xlabel("Time (model 1)")
ax[0].set_ylabel("Infections")
plt.show()